Hi, Mrtrixers,
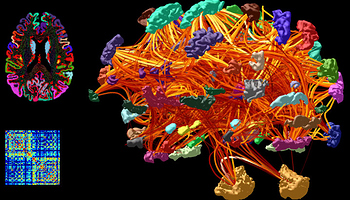
I intend to create volumes of cortical parcellation labels with streamlines assignment to parcellations. ( similar to Fig 1)
I firstly construct structural connectome with FreeSurfer’s aparc+aseg.mgz. then generate cortical labels volume with commands: mri_annotation2label and mri_label2vol. finally, I transform the label.nii.gz file to label.raw file(RW2 (Panasonic) ), so that I can render the labels.
Does the process make sense? The commands are as below.
Also, two another questions:
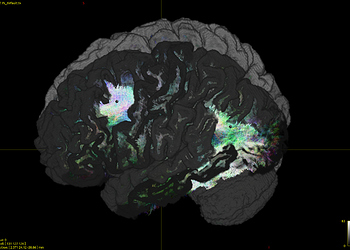
- as shown in Fig 2, It is clear that the nodes are there, but missing some cortical label surface when I
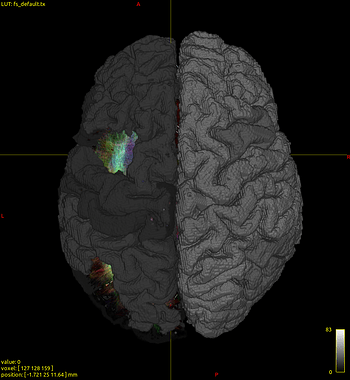
mrviewthe connectome node image. - as shown in Fig 3, lh.bankssts.labels does not show up when I choose “volume render” option.
Can anyone provide me some comments?
Thanks a lot!
Chaoqing
Fig 1 Structural Connectomics - Mrtrix3 homepage
Fig 2 mrview aparc+aseg_Desikan_Killany_nodes.nii
Fig 3
(ortho view) - lh.bankssts.label.nii.gz
(volume render) - lh.bankssts.label.nii.gz
Commands:
recon-all -i T1_source.nii.gz -s T1_source_bert -all -openmp 8
labelconvert T1_source_bert/mri/aparc+aseg.mgz /usr/local/freesurfer/FreeSurferColorLUT.txt …/…/share/mrtrix3/labelconvert/fs_default.txt Connectome/aparc+aseg_Desikan_Killany_nodes.nii
tck2connectome Output_ACT/DTI_processed_ACT_5TTwmmask_seedgmwmi_0.5M.tck Connectome/aparc+aseg_Desikan_Killany_nodes.nii Connectome/connectome.csv -out_assignments Connectome/assignments.csv
mkdir Connectome/Region2Region_tck
connectome2tck Output_ACT/DTI_processed_ACT_5TTwmmask_seedgmwmi_0.5M.tck > Connectome/assignments.csv Connectome/Region2Region_tck/edge_tracks_ -nodes 1 -files per_edge
mri_annotation2label --subject T1_source_bert/ --hemi lh --outdir /Volume/CorticalLabels
mri_annotation2label --subject T1_source_bert/ --hemi rh --outdir /Volume/CorticalLabelstkregister2 --mov T1_source_bert/mri/rawavg.mgz --noedit --s T1_source_bert/ --regheader --reg /Volume/CorticalLabels_register.dat
mri_label2vol --label /Volume/CorticalLabels/lh.bankssts.label --temp T1_source_bert/mri/rawavg.mgz --subject T1_source_bert/ --hemi lh --o /Volume/CorticalLabels_Volume/lh.bankssts.nii.gz --proj frac 0 1 .1 --fillthresh .3 --reg /Volume/CorticalLabels_register.dat
mri_label2vol --label /Volume/CorticalLabels/rh.bankssts.label --temp T1_source_bert/mri/rawavg.mgz --subject T1_source_bert/ --hemi rh --o /Volume/CorticalLabels_Volume/rh.bankssts.nii.gz --proj frac 0 1 .1 --fillthresh .3 --reg /Volume/CorticalLabels_register.dat