Cool! Thanks David!
Hi Marica_Padula,
This is a silly question. I would like to know how you generate this 3D rendering of the response function. Is it an in-built MrTrix function? or do you use matlab or python or some other external software?
I appreciate your help.
Thanks!
Julio Villalon
The command is shview. We should probably update the basic DWI tutorial to mention that…
Hi David,
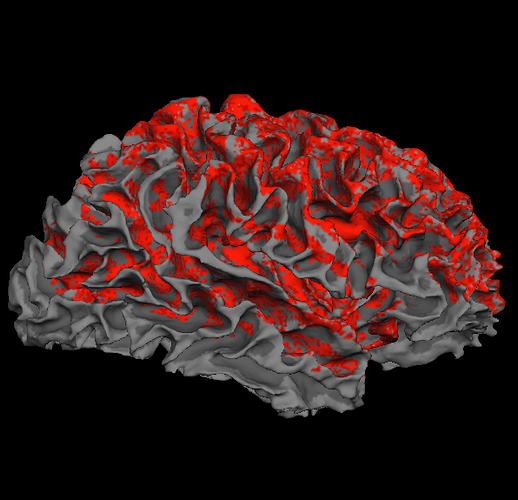
I actually tried that and it didn’t work. I used mesh_vertex_nearest to attribute each fibres termination to a vertex. If I plot the resulting vertices on the cortical surface I obtain what you can see in the screenshot. It seems evident to me that there is a problem of registration. I registered the DTI to the T1 image before performing the tractography, so my guess is that the coordinates in mm of mrtrix do not correspond to the coordinates of the vertices in the lh.white surface. Do you agree?
.