Dear Mrtrix experts,
We are preparing a DWI protocol for a 20-channel coil on a Siemens trio scanner. The protocol consists of a multi-shell (b0 b1000, and b2000) with both shells having 98 directions acquired in AP-PA directions. The voxel size is 1.7mm. I have two datasets acquired with 64-channel and 20 -channel coil, both with the same acquisition parameters. The 64-channel coil is the regular coil we use in our study, and 20 channel coil is for exceptional cases. Our primary purpose is tractography between the cerebellum and thalamus. Currently, I am inspecting the quality of data for 20 channel coil. As some quality checks, I tried to calculate SNR with methods @jdtournier explained in this thread:
I used the below commands to extract the wm mask from the largest b-value with 98 volumes, mean of signal in b0 b1000 b2000 after denoising, mean of the noise, and then divided the later values to get SNR within wm.
dwiextract dwi.mif -no_bzero -singleshell - | amp2sh - - | sh2power - -spectrum - | mrconvert - -coord 3 1 - | mrthreshold - wm_mask.mif
mrstats -output mean -mask wm_mask.mif noise.mif
dwiextract -shell X dwi_denoised.mif - | mrstats - output mean -mask wm_mask.mif -allvolumes -
for HARDI -20ch coil :
noise within WM : 367.762
signal within WM: b0=4328.56 b1000= 1955.16 b2000=1450.61
SNR _b0 = 11.77
SNR_b1000 = 5.31
SNR_b2000 = 3.94
for HARDI- 64ch coil
noise within WM : 221.267
Signal within WM: b0 = 3991.04, b1000=2118.96, b2000, 1324.15
SNR_b0 = 18.03
SNR_b1000 = 9.57
SNR_b2000 = 5.98
Is the notion of SNR>15 in b=0 shell and >5 in the outer shell standstill when calculating SNR within the wm, or it applies to the whole brain? How can I interpret these values if they are satisfactory for our purpose?
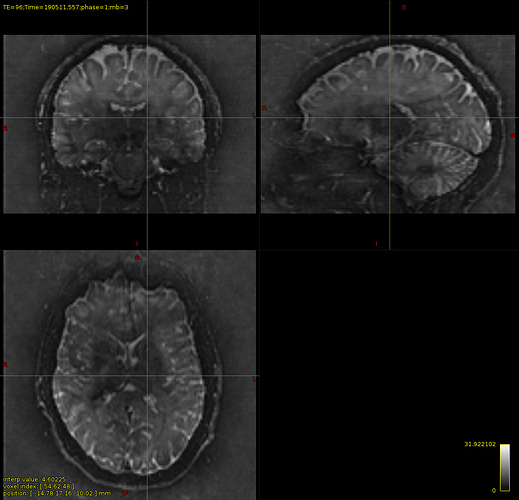
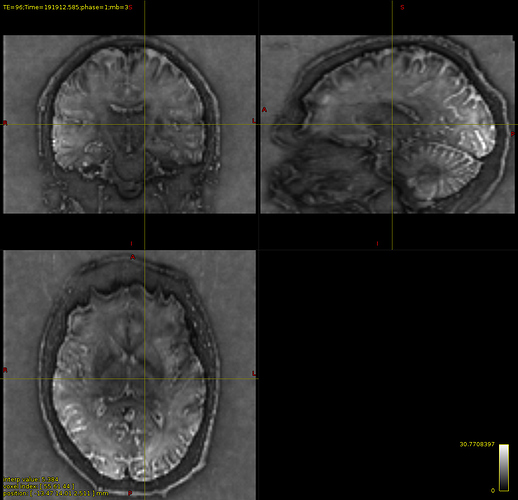
Furthermore, I generated the SNR map for both protocols, and the SNR seems to be lower in deep parts of the brain than the 64channel coil. However, I am not convinced if the 64 channel coil is good enough to take it as a reference for comparison.
Do you have any comments on our data quality, especially the one with 20 channel coil?
SNR for 20-channel coil
SNR for 64 channel coil
As the last question, how do you generally asses the quality of data? Except SNR and FODs, are there other considerations to take into account before commencing huge data collection?
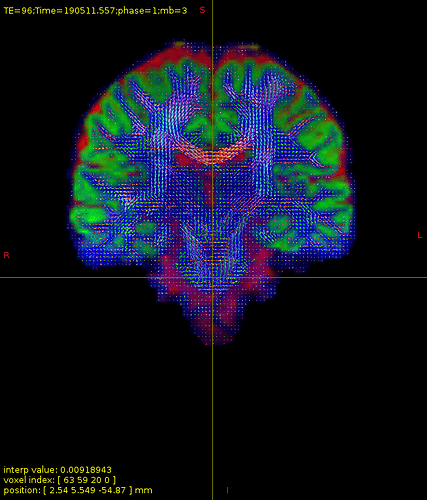
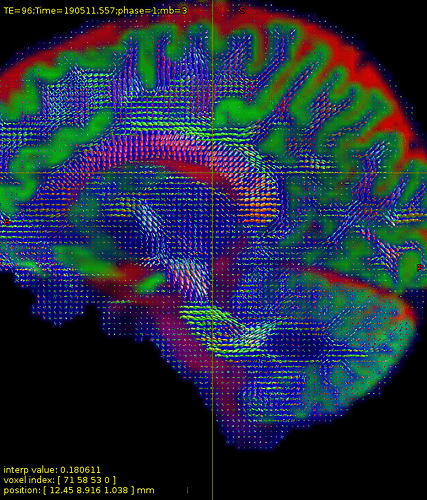
I have attached some screenshots(20ch coil) of FODs which might be helpful as further inspection.
Thanks!