Hi all,
I am following Andy’s Brain Book MRtrix tutorial #7: Streamlines (MRtrix Tutorial #7: Streamlines — Andy's Brain Book 1.0 documentation)
and was trying to do a deterministic tractography by FACT algorithm.
I couldn’t find a standard explanation on conducting deterministic tractography,
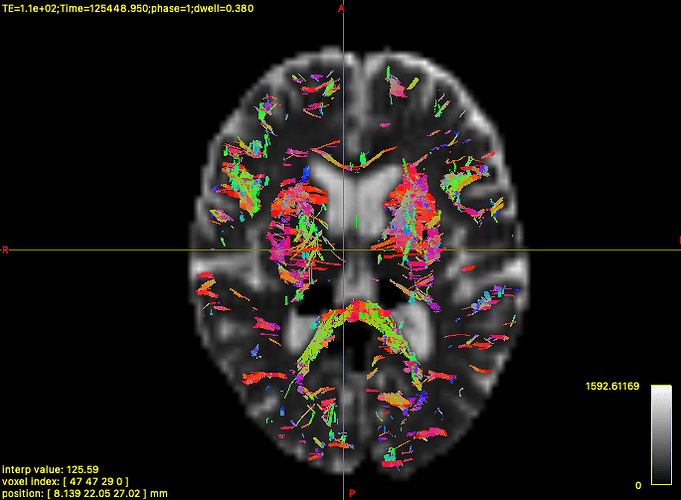
I got stuck on a problem that streamlines are not connected like the following picture.
original code:
tckgen -act 5tt_coreg.mif -backtrack -seed_gmwmi gmwmSeed_coreg.mif -nthreads 8 -maxlength 250 -cutoff 0.06 -select 10000000 wmfod_norm.mif tracks_10M.tck
the code I wrote:
tckgen -act 5tt_coreg.mif -seed_gmwmi gmwmSeed_coreg.mif -nthreads 8 -maxlength 250 -cutoff 0.06 -select 10000000 wmfod_norm.mif tracks_10M_det.tck -algorithm FACT
Was there anything wrong(perhaps maxlength or cutoff…?) with the code above?
Can you tell me the general settings of deterministic tractography?
I want to find out what I had missed, but cannot find the right answer.
Thank you so much for your help!
Hope you have a great day.
Best wishes,
Sujin.