Hi Michelle,
First up:
The group template mask consisted of regions that were not part of the brain in all subjects due to differences in FOV.
Normally the “group template mask” is defined as the regions that contain valid image data for all subjects; so presumably you must have derived that mask in some other fashion?
Regardless, I’ll draw your attention to this potential solution if the variance in coverage is an issue.
In the first approach, we mapped the scalar value in each voxel to all fixels within that voxel (voxel2fixel of the template mask) and then multiplied the template mask with the mask containing a reduced number of streamlines (150) using mrcalc. We used the adjusted 150-streamlines mask to perform fixelcfestats in the regions that were present in all included subjects.
I think there’s a couple of details missing in here. Firstly, given you said you’re performing an FBA, it’s not clear what “the scalar value” is. Is this referring to the ones and zeroes of a template voxel mask (however that was derived)? Secondly, I presume that you’re using tck2fixel to get a streamline count per fixel and applying a threshold of 150 to produce a fixel mask, but it’s not immediately clear with what information this is being combined; my best guess is that it is the template voxel mask that has been manually edited and then projected to fixels using voxel2fixel. I’m actually a big fan of using thresholds on multiple different sources of information in order to define the processing mask for fixelcfestats; it was just a little difficult to follow here exactly what was being done to what data.
In the second approach, we used the adjusted template mask from the white matter template analysis fixel mask onwards.
I think what you’re referring to here is that, instead of simply deriving a fixel mask to be applied solely during the fixelcfestats call, you have instead performed your manual manipulation of the template voxel mask at the point in the documented pipeline where that mask is derived, and then utilised that mask throughout the remainder of following that specific pipeline step-by-step?
the results were completely different.
Assuming that my interpretations of your descriptions are correct, and those are in turn faithful to what was actually performed, the key difference between the two is that in the former case, fixels that are eventually excluded from statistical inference are nevertheless included in the fixel-fixel connectivity matrix, and thus contribute to fixel data smoothing, whereas in the latter those fixels are never even segmented in the first place. One thing that’s been playing on my mind ahead of revising recommendations for the default pipeline (#2273) is that it’s not completely obvious whether or not the same fixel mask should be used for data smoothing as is to be used for statistical inference; they don’t have to be the same mask given the separation of these steps into different commands that happened in version 3.0.0.
But having said that, I wouldn’t expect a subtle change in smoothing mask to produce the kind of drastic change in outcome you’re describing. So maybe we should look for more substantial issues.
For instance, if you have used a fixel-fixel connectivity matrix computed from one set of fixels to smooth & perform statistical inference on a different set of fixels (which is what you have if you’ve provided a different mask to fod2fixel between the two approaches), I don’t recall whether an outright error will be produced (it probably should be), but if there isn’t, that’d completely bork your statistical power, since it would be comparable to filling your fixel-fixel connectivity matrix with noise.
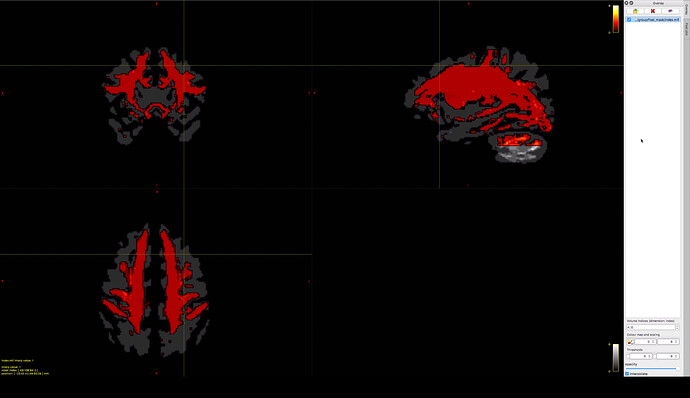
If we overlay the index-file of the newly created fixel mask on the fixel mask of the first approach,
I wouldn’t normally think of a fixel mask as “having” an index image; I prefer to think of a fixel set being defined by the index image, and a fixel mask is simply a bitwise fixel data file that accompanies that fixel set. The background image also appears to be a fixel index image, not a mask.
The screenshot is slightly visually deceiving because of the combination of interpolation and no transparency on the overlay, which together produces that black halo around the overlay image and kinda makes it look in places like the background image contains only the periphery and not the WM core.
(the yellow cross shows the significant region of the first approach)
Obviously if the region of interest is absent from the revised processing mask, it will be impossible to attribute statistical significance to such. But I don’t know how specific the indication of that location is from just a focus point without the p<0.05 region actually being shown.
If we did the analysis without adjusting the group template mask, the results were quite similar to the first approach, …
This I’m finding slightly difficult to interpret. What you’re referring to is not the first approach (i.e. using streamline count and presence of data for all subjects to define a mask for fixelcfestats only), since that’s what you’re contrasting against. So presumably you’re talking about using the template mask throughout the rest of the pipeline, as per the documentation, but without having performed any manual editing of the mask? So this is almost a “zero-th approach”, with no streamline density threshold and no manual mask editing. In that case, I wouldn’t expect the results to differ massively from your “first approach”; it’s the stark discrepancy between the “first” and “second” approaches that needs to be interrogated more closely.
… except for the fact that the regions that weren’t present in all the subjects, were significant as well.
Supposing that in voxels where image coverage for a particular subject was not achieved, the DWI data are zero-filled (which is what eddy does if DWI data for any volume for a particular voxel would need to come from outside the FoV), then if there is any correlation between your effect of interest and a reduction in DWI coverage (e.g. a pathological cohort that moves more), then those zero values will provide very strong numerical evidence for your hypothesis. Indeed even if you were to accurately mask in template space based on the presence of image data in subject space, the presence of that zero-filling adjacent to such a mask can influence the derivation of interpolated values within the intersection mask in template space.
I hope there’s something resembling a clue for a solution in there 
Rob