Hi everyone,
I want to use MRtrix in order to create a connectome with number of streamlines connecting ROIs of one specific atlas.
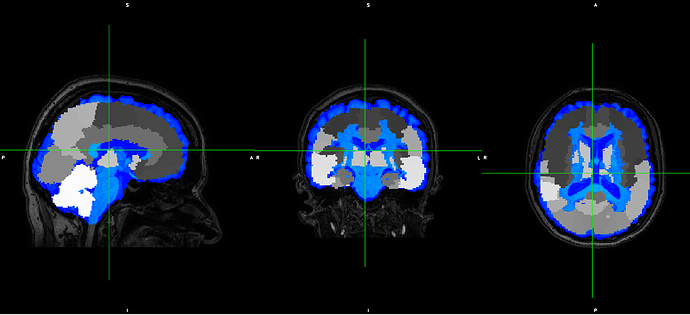
Following the instructions (Structural connectome construction — MRtrix3 3.0 documentation) I created the first connectome using the default Freesurfer’s recon-all output as parcellized brain (aparc+aseg). However, I need to change the default options, creating a connectome setting a different atlas for GM segmentation and consequently for connectome nodes.
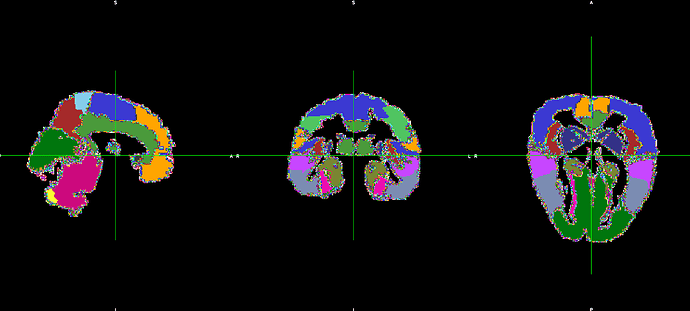
I tried for AAL atlas following different discussions (e.g. Problems with AAL parcellation - #2 by rsmith). These are the commands I run
flirt -in subj01_T1_bet.nii.gz -ref ROI_MNI_V4.nii -out T12AAL_nn.nii -omat T12AAL_nn.mat -interp nearestneighbour -dof 12
convert_xfm -inverse T12AAL_nn.mat -omat AAL2T1_nn.mat
flirt -in ROI_MNI_V4.nii -ref subj_T1_bet.nii.gz -interp nearestneighbour -applyxfm -init AAL2T1_nn.mat -out AAL2T1_nn.nii
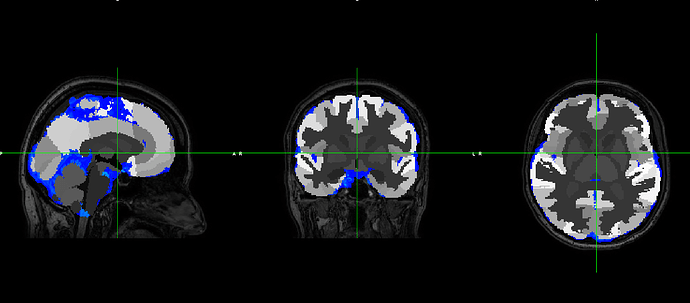
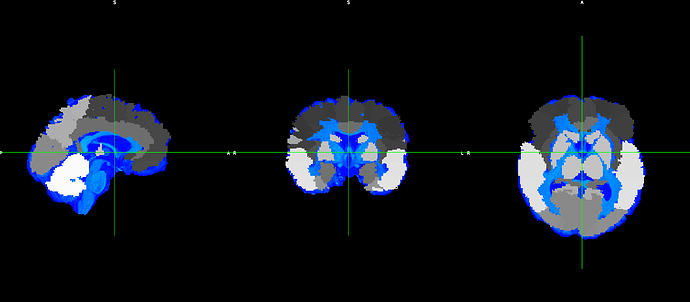
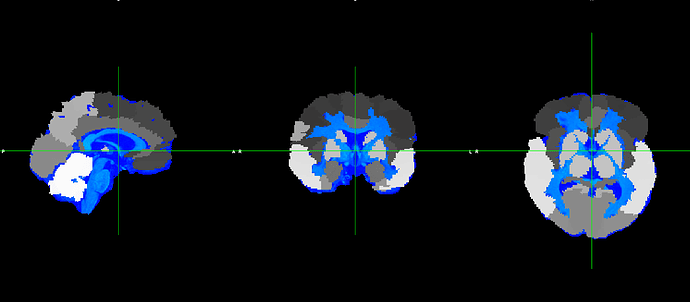
However, the output does not fit with the original T1
Are the steps correct in general?
Then, what can I do for obtaining a good T1 parcellisation based on an atlas different from the Freesurfer’s one?
Thanks