Dear experts,
I am processing ADNI data and performing EPI distortion correction using epi_reg as it has fieldmap only in the form of phase/mag scans. I used mrconvert to convert DWI dicoms to .mif along with its -json_export flag. In order to find phase encoding direction I did mrinfo of extracted DWI image and also compared it with exported json file. Unfortunately, I see different polarity of phase encoding direction, please see below.
mrinfo DWI.mif
************************************************
Image: "DWI.mif"
************************************************
Dimensions: 116 x 116 x 80 x 55
Voxel size: 2 x 2 x 2 x ?
Data strides: [ -1 -2 3 4 ]
Format: MRtrix
Data type: unsigned 16 bit integer (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0 -0 -115.4
0 1 -0 -101.8
-0 -0 1 -86.85
EchoTime: 0.056
FlipAngle: 90
MultibandAccelerationFactor: 1
PhaseEncodingDirection: j-
RepetitionTime: 7.2
SliceEncodingDirection: k
SliceTiming: 3.58999991,0,3.68000007,0.0900000036,3.76749992,0.180000007,3.85750008,0.267500013,3.94749999,0.357499987,4.0374999,0.44749999,4.12750006,0.537500024,4.21750021,0.627499998,4.30749989,0.717499971,4.39750004,0.807500005,4.48750019,0.897499979,4.57749987,0.987500012,4.66499996,1.07749999,4.75500011,1.16499996,4.84499979,1.255,4.93499994,1.34500003,5.0250001,1.43499994,5.11499977,1.52499998,5.20499992,1.61500001,5.29500008,1.70500004,5.38500023,1.79499996,5.47249985,1.88499999,5.5625,1.97500002,5.65250015,2.0625,5.74249983,2.15249991,5.83249998,2.24250007,5.92250013,2.33249998,6.01249981,2.4224999,6.10249996,2.51250005,6.19250011,2.60249996,6.28249979,2.69250011,6.36999989,2.78250003,6.46000004,2.86999989,6.55000019,2.96000004,6.63999987,3.04999995,6.73000002,3.1400001,6.82000017,3.23000002,6.90999985,3.31999993,7,3.41000009,7.09000015,3.5
TotalReadoutTime: 0.0336
command_history: mrconvert S574073 DWI.mif -json_export DWI_mif.json -export_pe_table pe_mif.txt (version=3Tissue_v5.2.8)
comments: 002_S_0413 (002_S_0413) [MR] Axial DTI
study: ADNI3 Basic Prisma ADNI3 Basic [ ORIGINAL PRIMARY DIFFUSION NONE ND NORM MOSAIC ]
DOB: 21/06/1930
DOS: 21/06/2017 13:48:54
dw_scheme: 0,0,0,0
[55 entries] 0,0,0,0
...
-0.070209229999999997,0.99701636999999999,0.032078339999999997,1000
0.42905900000000002,0.76815146000000001,-0.47523843999999998,1000
mrtrix_version: 3Tissue_v5.2.8
cat DWI_mif.json
************************************************
{
"EchoTime": "0.056",
"FlipAngle": "90",
"MultibandAccelerationFactor": "1",
"PhaseEncodingDirection": "j",
"RepetitionTime": "7.2",
"SliceEncodingDirection": "k",
"SliceTiming": "3.58999991,0,3.68000007,0.0900000036,3.76749992,0.180000007,3.85750008,0.267500013,3.94749999,0.357499987,4.0374999,0.44749999,4.12750006,0.537500024,4.21750021,0.627499998,4.30749989,0.717499971,4.39750004,0.807500005,4.48750019,0.897499979,4.57749987,0.987500012,4.66499996,1.07749999,4.75500011,1.16499996,4.84499979,1.255,4.93499994,1.34500003,5.0250001,1.43499994,5.11499977,1.52499998,5.20499992,1.61500001,5.29500008,1.70500004,5.38500023,1.79499996,5.47249985,1.88499999,5.5625,1.97500002,5.65250015,2.0625,5.74249983,2.15249991,5.83249998,2.24250007,5.92250013,2.33249998,6.01249981,2.4224999,6.10249996,2.51250005,6.19250011,2.60249996,6.28249979,2.69250011,6.36999989,2.78250003,6.46000004,2.86999989,6.55000019,2.96000004,6.63999987,3.04999995,6.73000002,3.1400001,6.82000017,3.23000002,6.90999985,3.31999993,7,3.41000009,7.09000015,3.5",
"TotalReadoutTime": "0.0336",
"comments": "002_S_0413 (002_S_0413) [MR] Axial DTI\nstudy: ADNI3 Basic Prisma ADNI3 Basic [ ORIGINAL PRIMARY DIFFUSION NONE ND NORM MOSAIC ]\nDOB: 21/06/1930\nDOS: 21/06/2017 13:48:54",
"dw_scheme": "0,0,0,0\n0,0,0,0\n0.43989815999999998,-0.33589560000000002,0.83286457999999997,1000\n-0.50804800000000006,-0.39628845000000001,-0.76474993999999996,1000\n0.14905657,-0.029167149999999999,-0.98839831,1000\n0,0,0,0\n0.56873286000000001,-0.71889256999999995,-0.39967024000000001,1000\n0.85194135000000004,-0.49273362999999998,0.17722699,1000\n0.14967976999999999,-0.42318463000000001,-0.89359421000000006,1000\n-0.39273444000000002,0.61961973000000004,0.67958145999999997,1000\n-0.73417931999999997,0.67150330999999996,0.10032070999999999,1000\n0.10245296,-0.76195592000000001,-0.63947385999999995,1000\n-0.18153580999999999,-0.23584774,-0.95468354,1000\n-0.0063826999999999998,0.79189503000000006,-0.61062395999999997,1000\n0.58592253999999999,0.77770852999999995,0.22773727999999999,1000\n-0.23924466999999999,0.91302335000000001,0.33037883000000001,1000\n0,0,0,0\n0.72041087999999998,-0.077146670000000001,-0.68924344000000004,1000\n-0.30029454999999999,-0.60058904000000002,0.74102354000000004,1000\n0.21523517,0.32601148000000002,-0.92053812999999995,1000\n0.15779425,0.58392465000000005,0.79632449000000005,1000\n0.35299735999999998,-0.68359548000000003,0.63881946000000001,1000\n-0.72240572999999997,0.13024057,-0.67909306000000003,1000\n0.78358406000000003,0.21103417999999999,0.58434635000000001,1000\n0.89609450000000002,-0.19314595000000001,0.39963660000000001,1000\n-0.23536652,0.91566581000000002,-0.32582044999999998,1000\n0.53192472000000002,0.83492535000000001,-0.14126427,1000\n0,0,0,0\n0.95507788999999998,0.26990809999999998,-0.12237621999999999,1000\n-0.46894637,0.26454781999999999,0.84267669999999995,1000\n-0.50867753999999998,-0.15774050000000001,0.84638363000000005,1000\n-0.82684857,-0.56102216000000005,-0.039691560000000001,1000\n-0.83448279000000003,-0.23042341,0.50054323999999994,1000\n-0.98982846999999996,0.12624163999999999,-0.065594780000000005,1000\n0.60907120000000003,0.43733569999999999,-0.66164171999999999,1000\n-0.73246217000000002,0.42271435000000002,0.53367739999999997,1000\n-0.94699842000000001,-0.20561542999999999,-0.24681243,1000\n0.89638494999999996,-0.38572007000000003,-0.21843525999999999,1000\n0,0,0,0\n0.74069297000000001,0.51961941,0.42587494999999997,1000\n0.45362595,-0.89113277000000002,-0.010290819999999999,1000\n0.12693903000000001,0.89855247999999999,0.42010713,1000\n-0.28302844999999999,-0.95019275000000003,-0.13049413000000001,1000\n-0.93593168000000004,0.07270625,0.34459498999999999,1000\n-0.089242269999999999,0.49946596999999998,-0.86172484999999999,1000\n-0.38259896999999998,-0.73187321000000005,-0.56389707,1000\n-0.66193073999999996,0.51412504999999997,-0.54545670999999996,1000\n0.58913243000000004,-0.75462454999999995,0.28890279000000002,1000\n-0.75377070999999995,-0.57402092000000005,0.31989020000000001,1000\n0.49238493999999999,0.055345419999999999,0.86861615999999997,1000\n0,0,0,0\n-0.14872740000000001,-0.95529717000000003,0.25551342999999999,1000\n0.16160287000000001,-0.14824474000000001,0.97565776000000004,1000\n-0.070209229999999997,0.99701636999999999,0.032078339999999997,1000\n0.42905900000000002,0.76815146000000001,-0.47523843999999998,1000"
}
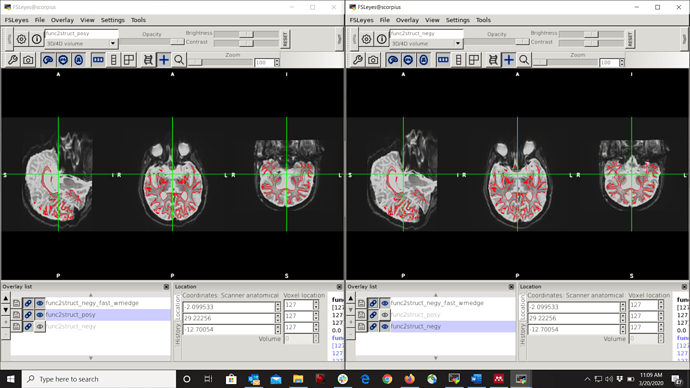
I also used dcm2niix to convert dicoms and compared the field with its extracted json file, which showed ‘-j’ as phase encoding direction similar to one exported by mrinfo. Besides, I performed EPI distortion correction with both positive and negative directions and reviewed unwarped images. Unwarped images look better when applying correction with +ve direction, please see attached screenshot.
What phase encoding direction should I use to correct for EPI and then for eddy correction using dwipreproc, and should I rely on mrinfo or exported json file?