Dear everyone,
I am currently post-processing DKI data using MRtrix. After some trial and error, I managed to use the development pathway to compute the kurtosis metrics. This is my first experience working with these types of metrics.
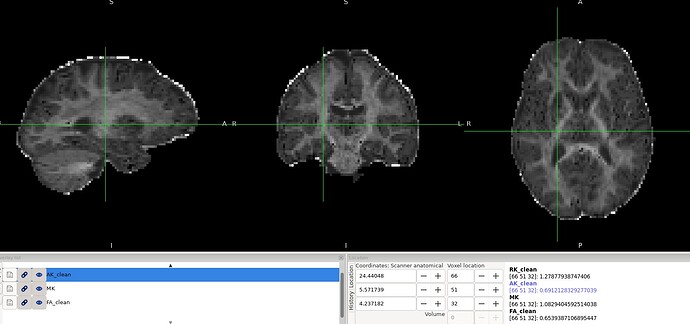
Overall, the results look reasonable; however, as shown in the image, there are a few black voxels, corresponding to values that are lower than expected, in the splenium and genu of the corpus callosum.
Are these voxels expected when computing kurtosis metrics? Is there a recommended way to mitigate or correct this issue? My main concern is that I plan to use DKI for tumor microstructure analysis, and I worry that similar low-value or black voxels may appear within the tumor region and affect interpretation.
Thank you very much for your time and help.