Dear all,
I’m trying to apply the MRtrix pipeline for connectome to my data. I have different dwi sequences. One of these is a single-shell one composed by 1 b0 and 40 b1000, no reverse encoding phase. For dwi2response I used dhollander algorithm using then dwi2fod msmt_csd with only wm and csf. After this I tried to register my data to the T1w in order to obtain the boundaries necessary for generating the streamlines. However, I am unsure of the outputs. These are the commands I run
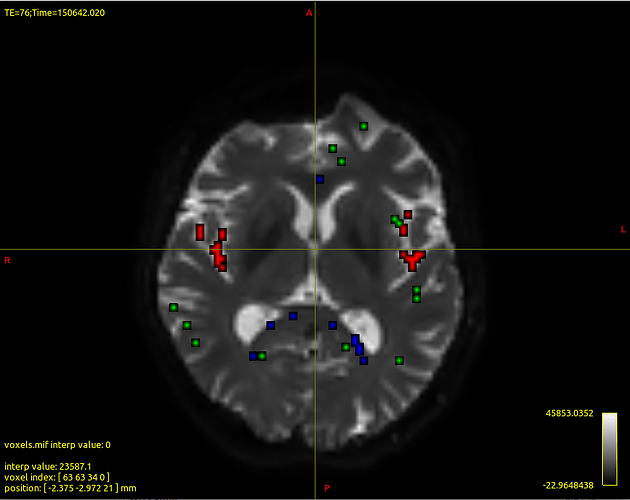
dwi2response dhollander subj01_PA_dwi_den_preproc_unbiased.mif wm_s.txt gm.txt csf.txt -voxels voxels_s.mif
dwi2fod msmt_csd subj01_PA_dwi_den_preproc_unbiased.mif -mask mask.mif wm.txt wmfod.mif csf.txt csffod.mif
mrconvert -coord 3 0 wmfod.mif - | mrcat csffod.mif - vf.mif
mtnormalise wmfod.mif wmfod_norm.mif csffod.mif csffod_norm.mif -mask mask.mif
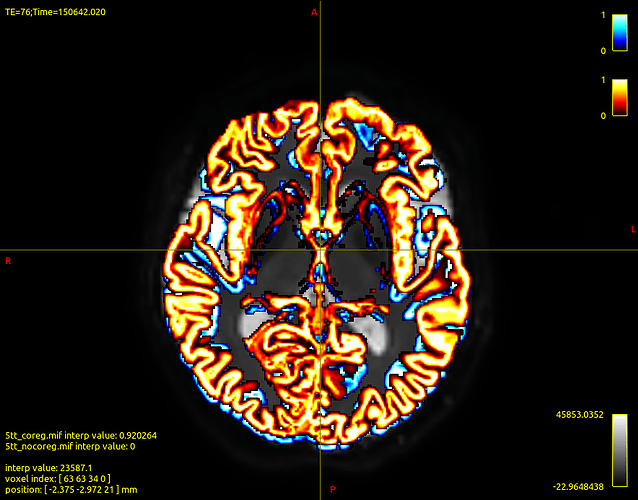
5ttgen fsl T1.mif 5tt_nocoreg.mif
Then I coregistered T1 with b0 of corrected dwi data and I created the boundaries
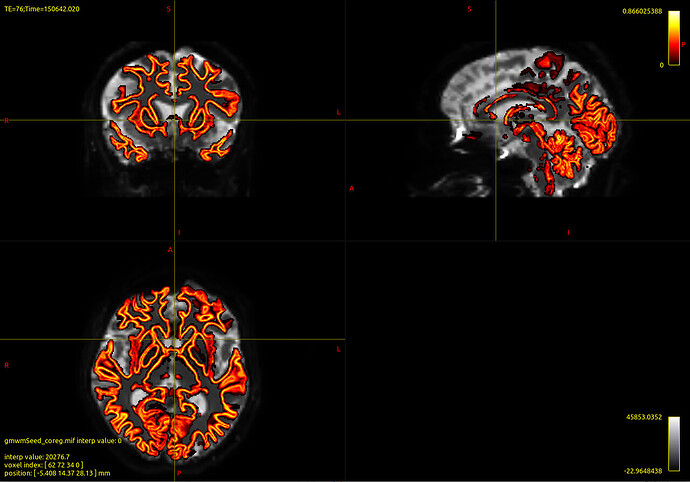
Attached the images output of dwi2response, registration and boundaries.
In dwi2response output voxels in ventricles were not considered among the ones used to model CSF diffusion. Is it correct?
Then, I don’t know if the registration went well. However, I would really appreciate feedbacks on this and on the boundaries created. Are they good? I think they are not so precise.
Many thanks
Beatrice