Hello,
I’m a bit confused about the relationship between the different softwares (FSL, MRTrix, DTI-TK) and storage formats for diffusion tensor images. Here’s what I would like to do:
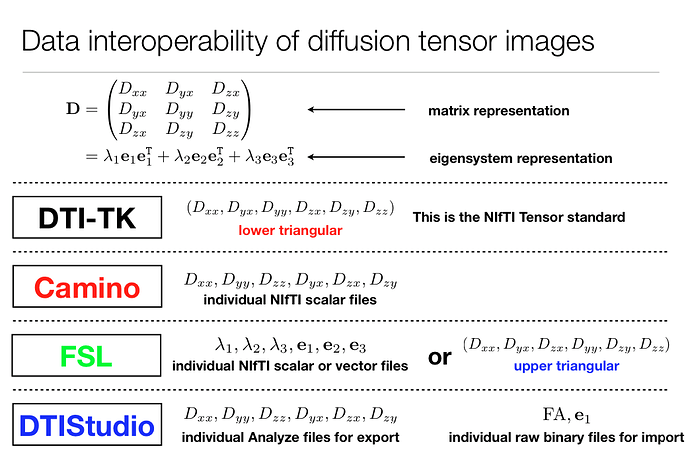
After running FDTs dtifit I want to use DTI-TK in order to register my DTI data to a white matter atlas (IIT). DTI offers a command (fsl_to_dtitk) for conversion of FSL´s tensor storage format (*_L1, *_V1, *_L2, *_V2, *_L3, *_V3 files) to so called NIfTI tensor format while also doing some other preprocessing such as transforming the origin to 0,0,0.
After running fsl_to_dtitk mrinfo yields:
Dimensions: 96 x 96 x 60 x 1 x 6
Voxel size: 2.5 x 2.5 x 2.30001 x 1 x 1
Data strides: [ 1 2 3 5 4 ]
Format: NIfTI-1.1 (GZip compressed)
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0 0 -0
0 1 0 -0
0 0 1 -0
While mrinfo on the FSL dtifit output *_tensor.nii.gz yields:
Dimensions: 96 x 96 x 60 x 6
Voxel size: 2.5 x 2.5 x 2.30001 x 7.53
Data strides: [ -1 2 3 4 ]
Format: NIfTI-1.1 (GZip compressed)
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0.002925 -0.003186 -120.9
-0.002675 0.9971 0.07601 -88.98
0.003399 -0.076 0.9971 -34.56
As I would like to extract diffusion metrics (FA, MD, …) from the tensor files after warping them to a common space I tried to use tensor2metric and compare the results on the 2 tensor images.
While extracting ADC (aka MD) seems to give the same result (just with a small translation), extracting FA only gives reasonable data on the FSL generated tensor image. Running tensor2metric -fa on the DTI-TK generated tensor image gives an image similar to a binarized brain mask.
DTI-TK also offers a command (TVEigenSystem) to bring the (warped) tensor image back to FSL format but that gives 6 files _e1, _e2, _e3, _lambda1, _lambda2, _lambda3. I’m not really sure in which order and how (maybe mrcat?) to bring them into one volume file which I can then use with mrtrix commands.
I also found this in the DTI-TK documentation:
From the dwi2tensor page I suppose MRTrix3 uses the same format as Camino:
The tensor coefficients are stored in the output image as follows:
volumes 0-5: D11, D22, D33, D12, D13, D23
DTI-TK offers a command to convert a Camino format stored DTI (in .hdr files) to the NIfTI tensor format but not the other way around.
I would be really glad if somehow had a short word of advice or a link where I could read up on this.
Thanks for reading,
Darius