Dear Mrtrix community,
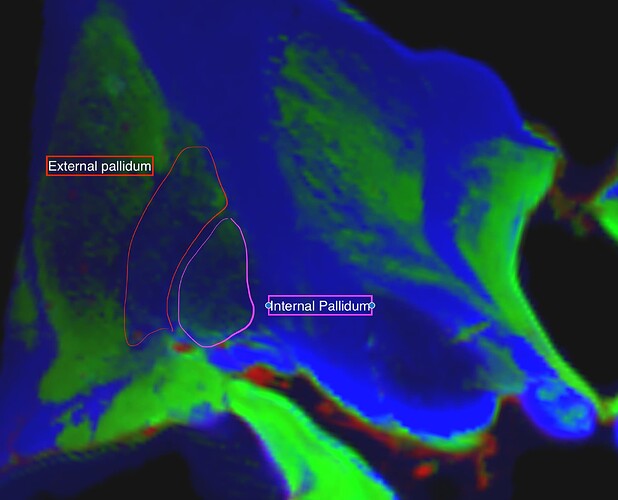
I am contacting you about the differences that I observe on the response functions of two different exvivo specimens in the same structures (Globus pallidus) in human.
The sequences were the same, 3 shells, 1000 4000 10000, 7 29 and 64 directions.
apart from the fact that we are in a very vague WM/GM context because I am interested in non-myelinated fibers, I think that the most desirable thing for the external pallidum is to be considered as white matter as it is is the case for H3H with rather blue pallidums. this being pased on microscopy studies and axonal tracing studies.
here are the maps that were created using
dwi2fod msmt_csd -force -nthreads 78 $Processed_data_folder/DWI_multishell_den_degibbs_preproc_bias_subtil.mif -mask $H4H_mask $Response_folder/response_wm_subtil.txt $FOD_folder/fod_wm_subtil.mif $Response_folder/response_gm_subtil.txt $FOD_folder/fod_gm_subtil.mif $Response_folder/response_csf_subtil.txt $FOD_folder/fod_csf_subtil.mif -nthreads 80 -debug
mrconvert -coord 3 0 \
$FOD_folder/fod_wm_subtil.mif - | mrcat $FOD_folder/fod_csf_subtil.mif \
$FOD_folder/fod_gm_subtil.mif - $FOD_folder/vfsubtil.mif -force -nthreads 80
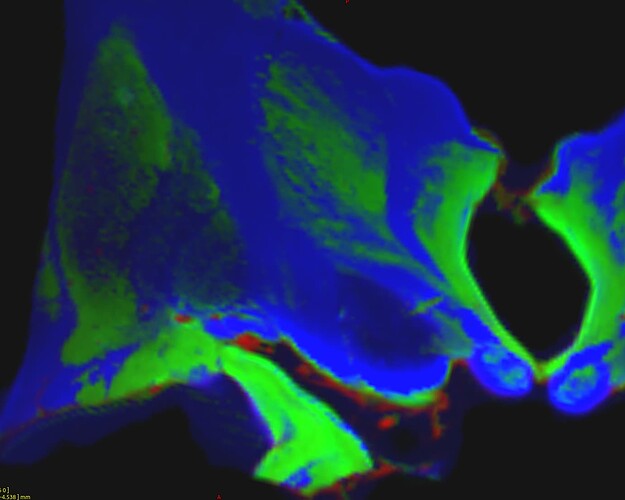
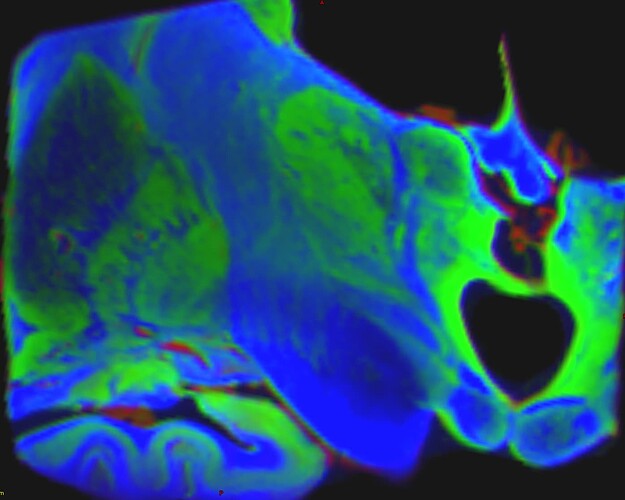
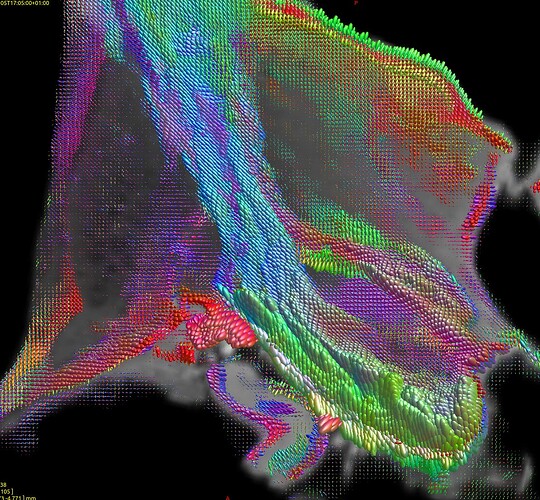
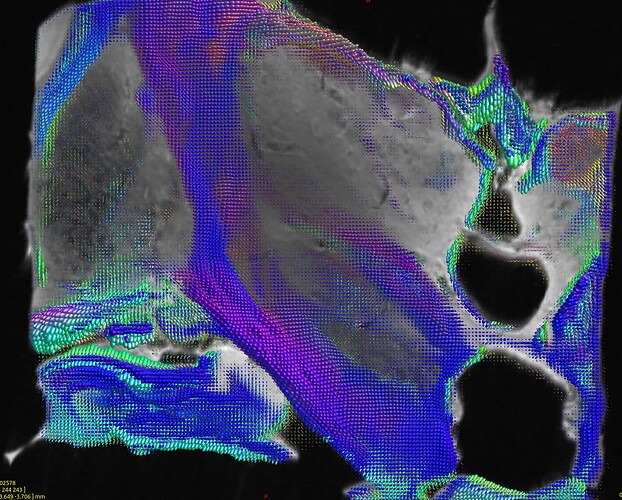
which gave thos FODs :
(do not take into account the color of the fods because the specimen were not placed exactly the same way in the scanner)
Where you can see much more fod signals in Pallidums in H3H wompared to H4H where the pallidums were considered as GM.
here is the code i used to create the image :
mrcat mifconverted_with_bvecbval/DWI_7dirs_B1000.mif mifconverted_with_bvecbval/DWI_29dirs_B4000.mif mifconverted_with_bvecbval/DWI_64dirs_B10000.mif processed_data/DWI_multishell2.mif (version=3.0.4-50-gf36f28c9)
dwidenoise processed_data/DWI_multishell.mif processed_data/DWI_multishell_den_subtil.mif -mask H4H_raw_datas/H4H_mask.nii -extent 7 -datatype float32 -noise processed_data/DWI_multishell_noise_subtil.mif -nthreads 80 -force -info -debug (version=3.0.4-50-gf36f28c9)
mrdegibbs processed_data/DWI_multishell_den_subtil.mif processed_data/DWI_multishell_den_degibbs_subtil.mif -nthreads 80 -debug (version=3.0.4-50-gf36f28c9)
/home/nicolas.tempier/mrtrix3/bin/dwifslpreproc processed_data/DWI_multishell_den_degibbs_subtil.mif processed_data/DWI_multishell_den_degibbs_preproc_subtil.mif -rpe_none -pe_dir AP -eddy_options ' --slm=quadratic --data_is_shelled --repol --residuals --cnr_maps --very_verbose --estimate_move_by_susceptibility' -eddyqc_all processed_data/eddy_qc_subtil -eddy_mask H4H_raw_datas/H4H_mask.nii -force -nthreads 80 (version=3.0.4-50-gf36f28c9)
/home/nicolas.tempier/mrtrix3/bin/dwibiascorrect ants processed_data/DWI_multishell_den_degibbs_preproc_subtil.mif processed_data/DWI_multishell_den_degibbs_preproc_bias_subtil.mif -mask processed_data/H4H_mask_aligned_subtil.nii -nthreads 80 -debug -nocleanup -bias processed_data/DWI_multishell_bias_subtil.mif (version=3.0.4-50-gf36f28c9)
dwi2fod msmt_csd -force -nthreads 78 processed_data/DWI_multishell_den_degibbs_preproc_bias_subtil.mif -mask H4H_raw_datas/H4H_mask.nii processed_data/responses/response_wm_subtil.txt processed_data/fod/fod_wm_subtil.mif processed_data/responses/response_gm_subtil.txt processed_data/fod/fod_gm_subtil.mif processed_data/responses/response_csf_subtil.txt processed_data/fod/fod_csf_subtil.mif -nthreads 80 -debug
Could I get your opinion on this please ?
Best,
Nicolas