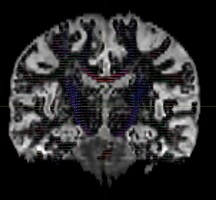
Hi all experts,
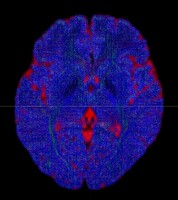
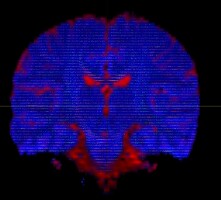
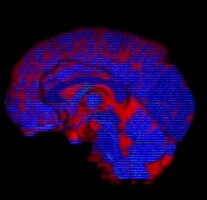
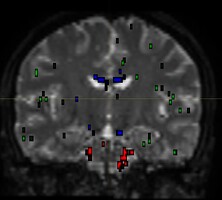
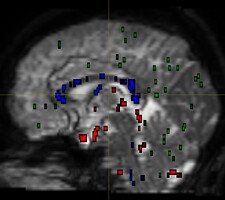
I am using dwi2fod msmt_csd to estimate fiber orientation distribution. It seems that white matter fibers are estimated in gray matter regions, as indicated by the substantial portion of blue region in the QC image.
dwi2fod QC images:
For your information:
- My data were acquired in one PE direction with two b-values only (0, 1000) – is msmt applicable to my data? If not, which option should I use instead?
- Outputs of dwi2response dhollander:
Any ideas of where do I have to look into?
Many thanks for your help,
Idy
Hi,
What exact command did you use for dwi2fod? Your data is “two shells” (0 and 1000). If you have only two shells you need to use the dwi2fod msmt with only two response functions, in this case the WM and the CSF. You won’t be able to do a three tissue diferentation with these algorithm with only two shells.
The alternative option is to look at the SS3T algorithm.
Best regards,
Manuel
1 Like
Hi Manuel,
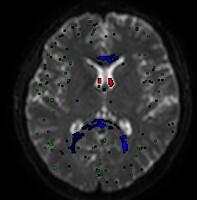
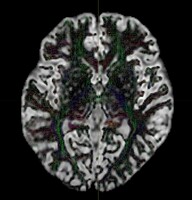
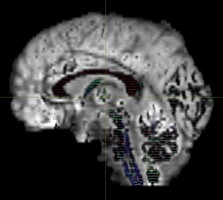
Thank you very much for your help!! I’ve re-run dwi2fod msmt with two RFs only (WM and GM) and it seems to produce correct results. The new output images:
This is my first time using MRtrix so I am a bit insecure of whether my results are correct. Although it looks reasonable to me that FOD were only estimated in the WM tracts as shown in the outputs, please do let me know in case it is still fallicious.
Many thanks!!!