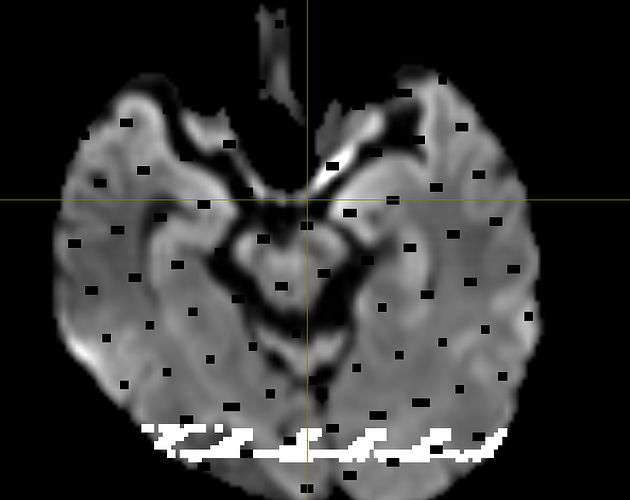
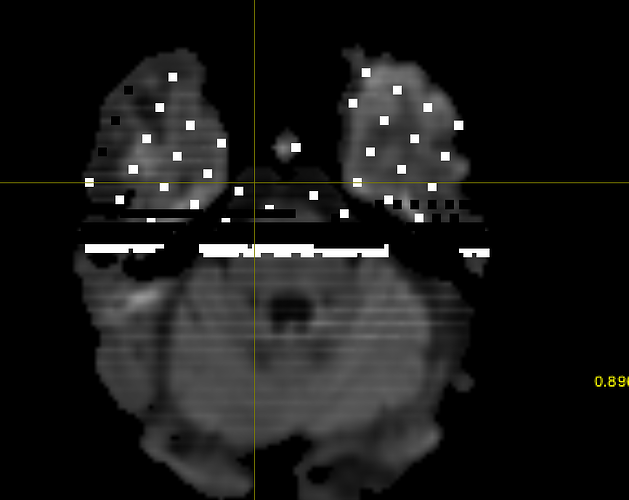
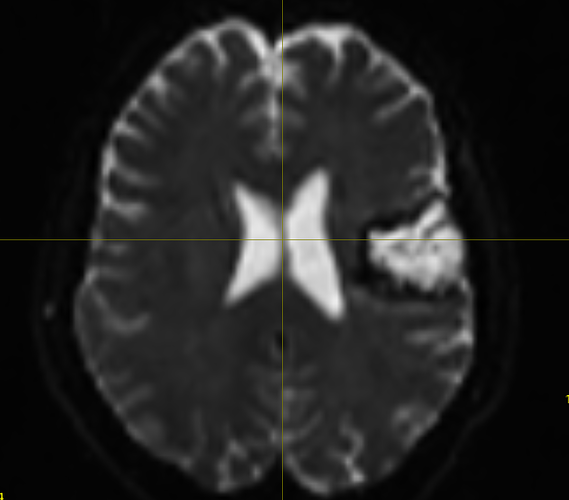
Hi! I’m running a normal msmt csd pipeline for some stroke patients, and while most participants are smooth, about 5/50 of them end up with wmfods that have a bunch missing voxels, or odd looking static. Their dwi_denoised_degibbs_preproc_unbiased_upsampled.mif files (what I am feeding into dwi2fod) alongside their masks are perfectly fine, which is what is rather confusing to me.
Here are screenshots of what I’m getting :
I’m wondering if that’s something unique to the participant that is causing the issue (lesion location/characteristics, or acquisition artifact for that individual)? Or if it’s something wrong that’s happening with the entire pipeline for all participants.
Thank you for the help!
That does not look right. If you run dwi2fod again on the same dataset, do you get exactly the same pattern?
Yup, rerunning it encounters the same issue. I’m using msmt_csd with 2 tissues (b=0, 1000).
The participants seem to be ones with a more noticeable lesion in their dwi image, so I’m thinking that could be the source of the distortion/error?
This is an example
Hi @bennnzhang,
The processing in dwi2fod is performed on a purely voxel-by-voxel basis. There is no way a distant lesion could give rise to these issues. My best guess is that there is some data corruption on one or more volumes of the input data, which isn’t immediately obvious as it might only affect one of the later volumes – you’d need to cycle through all of your volumes and check each one independently.
As a quick check, you could try to find any non-finite values (NaN or Inf) in your input dataset, using a command like this one:
mrcalc input_dwi.mif -finite -not - | mrmath - sum -axis 3 - | mrview -
This creates a binary image with 1s where the values are not finite, then compute the sum of that across volumes, and display it in the viewer. You’ll be looking at an image where the intensity in each voxel is the number of non-finite values across all volumes at that location. If you see non-zero values in the same places where you see corrupt values in the dwi2fod output, that means your input is already corrupt – nothing to do with dwi2fod. You’d then need to back-track through the steps and identify which command might have been responsible for this.
This isn’t fool-proof, by the way – we’re only checking for non-finite values. The data could be corrupted in other, less obvious way (e.g. just nonsense values, but not NaN or Inf as such). You’d still need to check each volume even if the command above doesn’t find any overtly corrupt voxels.
Personally, given the way the artefact manifests, I would suspect a hardware issue. Either faulty RAM, or a faulty hard drive. But you’ll first need to verify whether the input is indeed corrupt…
Cheers,
Donald.
2 Likes
Thanks so much for this detailed response!
I saw non-finite voxels in the upsampled image, but the non-upsampled image was perfectly fine, so it seems like somewhere in the upsampling process there was some corruption.
This makes sense as I had some trouble running mrgrid on the data directly on an external SSD, and will definitely have to go back and take a look at that process. I’m guessing your intuition regarding the hardware is correct.
I really appreciate your help. This has been stumping me for a pretty long time now. Cheers!
1 Like
To follow up on this, it was indeed an issue with the external drive I was using! I ran everything on my computer’s native hard drive and the wmfods turned out perfectly.
Appreciate your help.
Best, Ben
1 Like