Hello,
The code below is part of my preprocessing pipeline, which I ran in Neurodesk using MRtrix3 (version 3.0.4). My DWI data is multi-shell, with 1 b=0, 64 b=1500, and 64 b=3000 volumes (PhaseEncodingDirection = j).
#!/bin/bash
cd “/data/subjects”
ml mrtrix3/3.0.4
ml fsl/6.0.7.16
ml ants/2.5.3
for_each * : dwidenoise IN/dwi.mif IN/dwi_denoised.mif -noise IN/noise.mif
for_each * : mrdegibbs IN/dwi_denoised.mif IN/dwi_denoised_unringed.mif -axes 0,1
for_each * : dwifslpreproc IN/dwi_denoised_unringed.mif IN/dwi_denoised_unringed_preproc.mif -rpe_none -pe_dir j -eddy_options " --repol --cnr_maps --slm=linear" -eddyqc_all eddyQC
for_each * : dwibiascorrect ants IN/dwi_denoised_unringed_preproc.mif IN/dwi_denoised_unringed_preproc_unbiased.mif -bias IN/bias.mif
for_each * : dwi2mask IN/dwi_denoised_unringed_preproc_unbiased.mif IN/dwi_mask_unbiased.mif
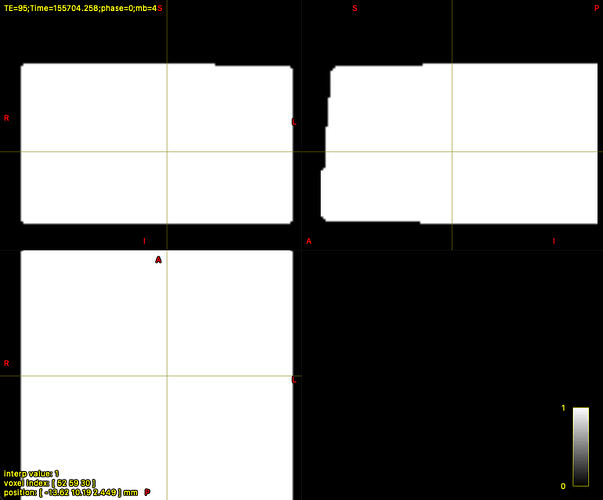
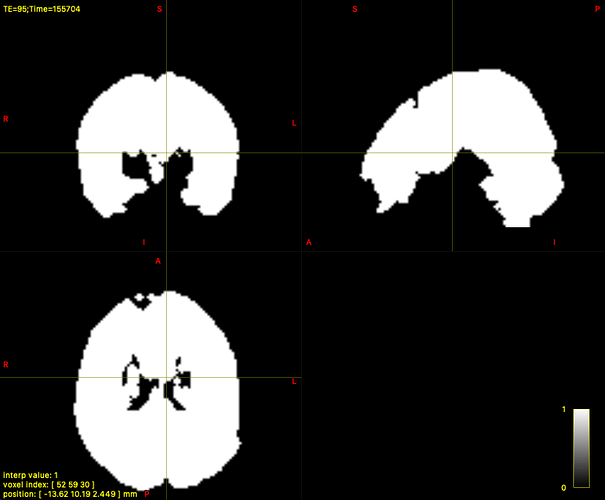
For most subjects, the dwi_mask_unbiased.mif was correctly generated. However, in 9 subjects, dwi2mask produces a completely white mask (i.e., the entire field of view is white instead of a brain-shaped mask).
Do you have any suggestions to improve the dwi2mask output in those cases?
Interestingly, in those 9 cases, dwifslpreproc creates a more accurate mask (eddy_mask.nii.gz) that appears to correctly segment the brain (but sometimes with holes).
Would it be appropriate to use eddy_mask.nii.gz as a replacement in these cases? If so, are there any references or best practices that support this?
Any advice is appreciated.
Thanks!
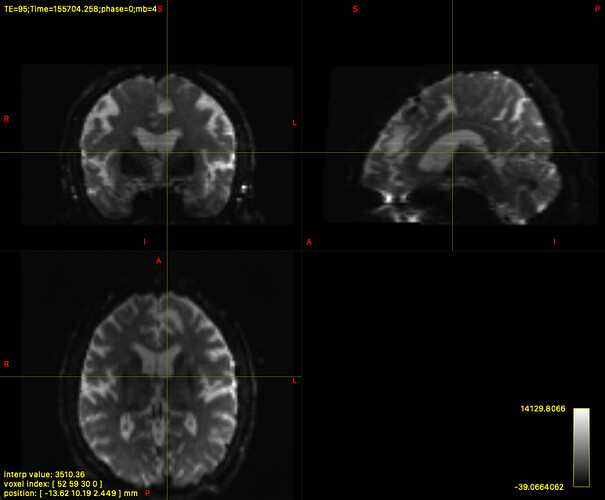
p.s. The following image is the dwi_denoised_unringed_preproc_unbiased.mif of the above masks: