Hi Mrtrix3 experts,
I have a multi-shell DWI data scan with 3 b-values including 0, 3000 and 6000. Can I go ahead using dwi2response dhollander to estimate response functions? Or should I need to extract them to single shell using dwi2extract and run dwi2response on every shells, then concatenate them at the end?
Thank you so much,
Van
Hi @vanlam2308,
Yep, 100% fine! That’s the very purpose of this algorithm even.
Nope! That would actually be wrong in certain ways; you’d get a multi-shell response in the end that’s a combination of single-shell responses from different sources (voxels in this case), something you definitely don’t want to have for various reasons.
dwi2response dhollander was designed for any number of b-values. The new 2019 version (which you’re probably not using here yet) even improves the single-fibre WM response estimation in particular for multiple b-values. (sources: ISMRM abstract and ISMRM talk)
If you’d like me to have a look just to be sure, happily post the contents of your estimated WM, GM and CSF response functions here. You can also get an image of the final voxel selection via the -voxels option. Here’s an old post on how you can visualise these voxels easily for inspection.
All the best,
Thijs
Hi Thijs,
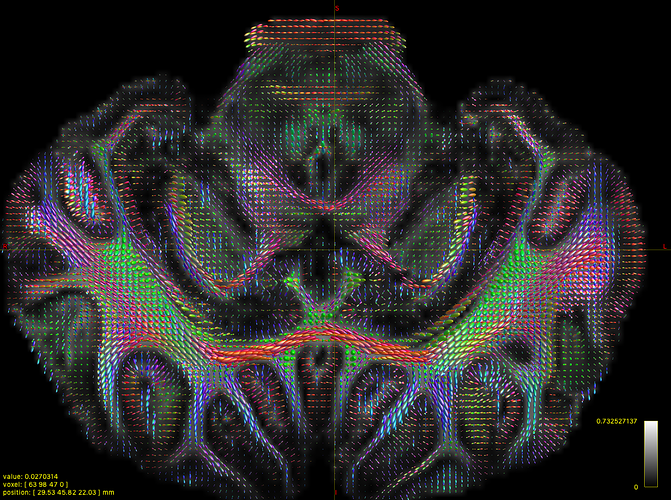
Thank you very much for your confirm. I have already generated WMFOD image. However, I found that there are many noise found in cortical regions. I do not know how to exclude them in fact. My data was generated using 3000 and 6000 b-value and this is ex-vivo scan on pig brain. I am attaching a screenshot below.
Wow, very nice dataset! Also, that does look quite good actually: I can clearly identify a lot of axonal projections into the cortex, as it should be  . If you’re still unsure, see my post above about posting an image showing the selected voxels that were used to calibrate the response functions. As your scan is ex-vivo, that’s sometimes a bit “special”. Not always a problem or even a challenge though… unless it is.
. If you’re still unsure, see my post above about posting an image showing the selected voxels that were used to calibrate the response functions. As your scan is ex-vivo, that’s sometimes a bit “special”. Not always a problem or even a challenge though… unless it is.  That explains your b=6000 choice though (which is a good one). The b=3000 should be regarded as a “low” b-value in this case (as opposed to in vivo scans where most would refer to b=3000 as a “high” b-value).
That explains your b=6000 choice though (which is a good one). The b=3000 should be regarded as a “low” b-value in this case (as opposed to in vivo scans where most would refer to b=3000 as a “high” b-value).
If you’re unsure about the WM FOD output itself, you can also show me the GM and CSF outputs from the 3-tissue CSD model as well (i.e. those that you get together with the WM FOD image). You should at least see some decent GM-like signal contribution across the entire cortex: that would then mean that such signal was indeed successfully filtered out of the WM FOD. If you’d like me to have a look at the dataset itself; always happy to when I find a bit of time: contact me privately in that case.
 . If you’re still unsure, see my post above about posting an image showing the selected voxels that were used to calibrate the response functions. As your scan is ex-vivo, that’s sometimes a bit “special”. Not always a problem or even a challenge though… unless it is.
. If you’re still unsure, see my post above about posting an image showing the selected voxels that were used to calibrate the response functions. As your scan is ex-vivo, that’s sometimes a bit “special”. Not always a problem or even a challenge though… unless it is.  That explains your b=6000 choice though (which is a good one). The b=3000 should be regarded as a “low” b-value in this case (as opposed to in vivo scans where most would refer to b=3000 as a “high” b-value).
That explains your b=6000 choice though (which is a good one). The b=3000 should be regarded as a “low” b-value in this case (as opposed to in vivo scans where most would refer to b=3000 as a “high” b-value).