Hi all,
I have single shell data with which I am using MRtrix 3Tissue_v5.2.9 to run multi-tissue CSD from the tutorial here: Fibre density and cross-section - Multi-tissue CSD — MRtrix 3.0 documentation. Information on my data is included here (image below)
Within my subjects directory are the individual subject folders (image below)
And within each subject folder are (image below) AP.mif.gz, PA.mif.gz, b0_pair.mif.gz and dwi.mif.gz
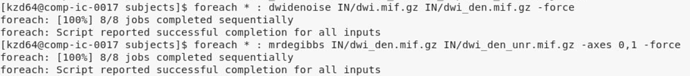
From the subjects directory, I am able to run denoising and unringing without issue (image below)
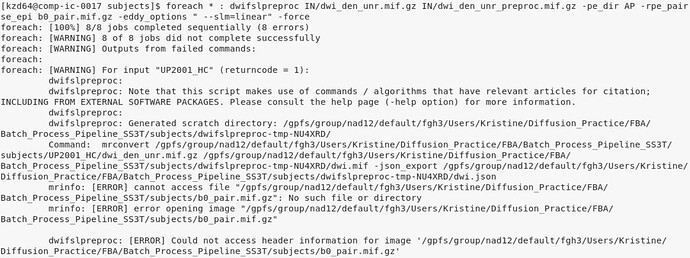
However, when running dwifslpreproc command after this, I receive the below error message for each of my 8 subjects (I included the " --slm=linear" option in the dwifslpreproc command since I previously received a message 'sampling of b=1000 shell is strongly asymmetric; distortion correction may benefit from use of: -eddy_options " … --slm=linear) on a previous run
Given MRtrix reports “no such file or directory”, I thought perhaps it was not looking in the subjects directory, but that does not appear to be the case. I then thought I had misspelled the b0_pair.mif.gz file either within each subject folder or the dwifslpreproc command, but I do not think that is it either. I then consulted the documentation for the command here dwifslpreproc — MRtrix 3.0 documentation, as well as some of the existing topics on dwifslpreproc here on the forum, but not sure what I am doing incorrectly in this step.
I will apologize if this has been answered in an existing question, learning how to implement this, troubleshoot, and post for the first time.
Cheers,
Kristine