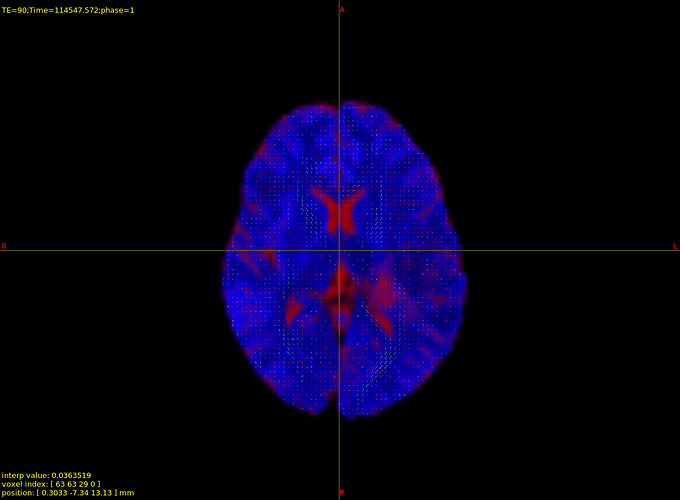
Hi Parand,
However, I see no change between my original and processed DTI images.
Rather than looking at the two images separately, or in two different windows, are they precisely numerically equivalent? You could also have a look at the motion / eddy current parameters estimated by eddy (they’ll be in the scratch directory if you run dwifslpreproc with the -nocleanup flag), to see if there is in fact genuinely zero correction being applied.
As far as I understood, there is nothing I can do about it at this point. I hope this effect doesn’t negatively impact my whole analysis.
SynB0-DISCO is worth attempting. I’m sure @schilkg1 would appreciate feedback on its operation in the presence of such lesions!
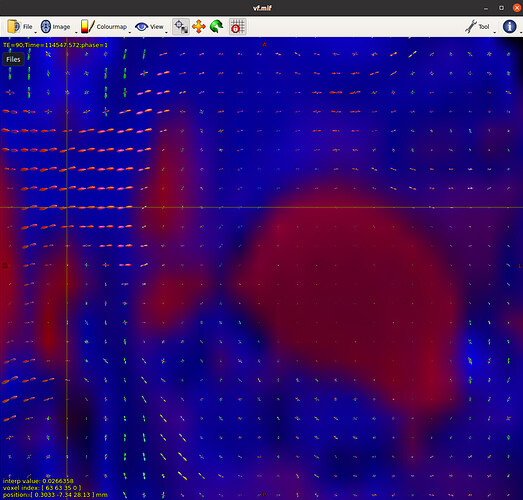
Now I am facing another barrier in Fiber Orientation Density and Normalization section.  /
/
When I look into vf.mif, I don’t see any gray matter information (green colour). I am not sure why!
I assume “vf.mif” is supposed to be “volume fractions”; not actually sure which document you are working off. But I’m guessing that you have single-shell data, in which case dwi2fod msmt_csd cannot estimate a three-tissue decomposition because there are only two unique b-values (admittedly it would likely be preferable for the command to generate an appropriate error message under such circumstances).
As you can see, there is a little bit of fiber tract information inside csf!
You can’t necessarily expect the WM ODF to be of exactly zero magnitude inside of regions that are otherwise considered to be CSF; there’s both the prospect of there actually being some restrictive structures in some such regions, and the possibility of image noise making the diffusion-weighted signal look like CSF with just a smidgen of WM rather than being a perfect match to the CSF response function. Indeed if you were to consider the limit where the CSF response function is based off of the most extreme exemplar CSF voxel, then every voxel other than that one exemplar voxel would have a non-zero WM ODF.
Also, when I run normalization command, I receive the following error: mtnormalise: [ERROR] Non-positive tissue balance factor was computed. Balance factors: -nan -nan -nan
This may be related to having provided the empty GM ODF to mtnormalise. If you re-run mtnormalise using only the two valid ODF images, and still run into this problem, there are a number of threads on the forum that go through the debugging process for this specific error message.
Cheers
Rob
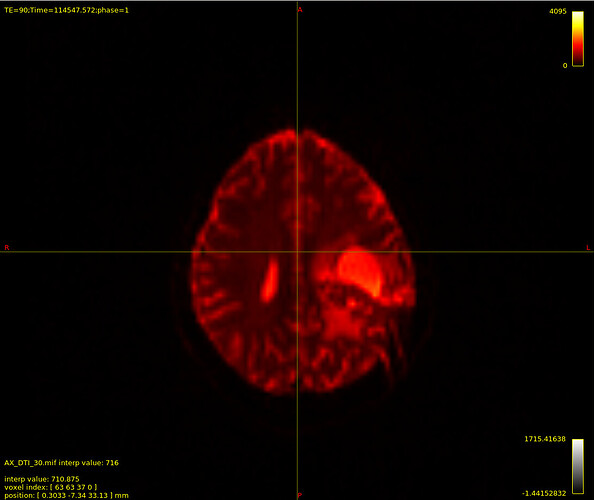
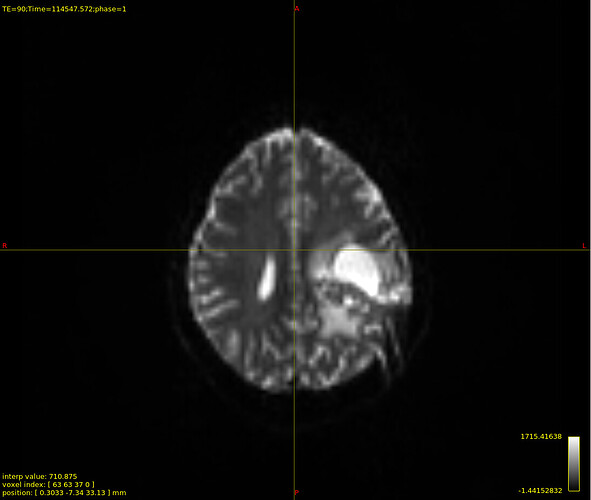
 /
/