Hi rob,
I tested this issue out again and find out following: as you mentioned, all approaches run into same warning resulting with no Eddy QC .pdf and .json report produced (sorry for mystification). I am sure it is not a problem of FSL installation since on a 3 different dataset I did not faced this problem.
my dataset:
-
b_value (No of dirs): 0 (11), 750 (17), 1500 (26), 3000 (35), 5000 (43)
-
full rPE: AP_PA with identical dw_schemes (checked all 12 decimal places)
-
Dimension 120x120x75x132
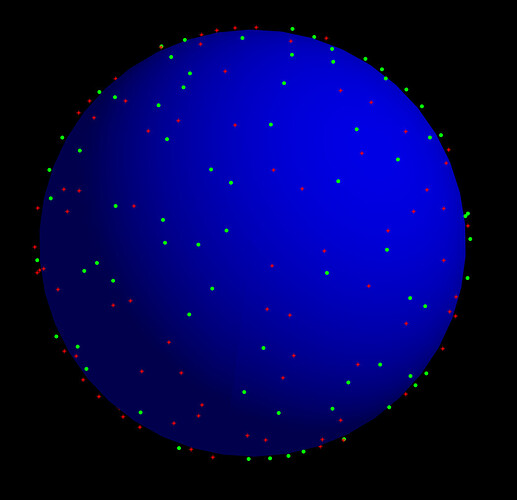
I am giving here a prtsc from Thijs Dhollander’s script for generating image of b vectors distribution:
When I run dirstat I’ve got following warning for each shell with value varying from 0.14 to 0.26
I applied on a data same pipeline as usual, ( mrconvert dcm to mif; no denoising since it produces problems when eddy called too - we discussed it in a separate thread; mrdeggibs, dwifslpreproc). For the last step I have run following command:
dwifslpreproc ${i}_dwi_den_unr_AP_PA.mif ${i}_dwi_den_unr_preproc.mif -pe_dir AP -rpe_all -nocleanup -eddy_options " --slm=linear --repol --mporder=16 --cnr_maps " -eddyqc_all ${Dir}/${i}/Mrtrix3_proc/QC
During the dwifslpreproc I’ve got warning about not providing report but I am not able to uncover why:
Versions tested (slspec files copied from -nocleanup):
- mrconvert .dcm to .mif, no slspec file provided
0 25 50
2 27 52
4 29 54
6 31 56
8 33 58
10 35 60
12 37 62
14 39 64
16 41 66
18 43 68
20 45 70
22 47 72
24 49 74 75
1 26 51
3 28 53
5 30 55
7 32 57
9 34 59
11 36 61
13 38 63
15 40 65
17 42 67
19 44 69
21 46 71
23 48 73
- mrconvert .dcm to .mif, slspec file provided
0 41 57
25 66 9
50 18 34
2 43 59
27 68 11
52 20 36
4 45 61
29 70 13
54 22 38
6 47 63
31 72 15
56 24 40
8 49 65
33 74 17 75
58 1 42
10 26 67
35 51 19
60 3 44
12 28 69
37 53 21
62 5 46
14 30 71
39 55 23
64 7 48
16 32 73
provided by me was:
0 41 57
25 66 9
50 18 34
2 43 59
27 68 11
52 20 36
4 45 61
29 70 13
54 22 38
6 47 63
31 72 15
56 24 40
8 49 65
33 74 17
58 1 42
10 26 67
35 51 19
60 3 44
12 28 69
37 53 21
62 5 46
14 30 71
39 55 23
64 7 48
16 32 73
- mrconvert .dcm to .nii to .mif (to have no info about slicetiming in a header of .mif), slspec file provided
0 41 57
25 66 9
50 18 34
2 43 59
27 68 11
52 20 36
4 45 61
29 70 13
54 22 38
6 47 63
31 72 15
56 24 40
8 49 65
33 74 17 75
58 1 42
10 26 67
35 51 19
60 3 44
12 28 69
37 53 21
62 5 46
14 30 71
39 55 23
64 7 48
16 32 73
Provided by me same as in a 2nd case.
Slspec files provided by me were generated with my own script. Original slicetiming from dicom header was as follows:
AP:
0.0000,1.6700,0.1300,1.7975,0.2575,1.9275,0.3850,2.0550,0.5150,2.1825,0.6425,2.3125,0.7725,2.4400,0.9000,2.5675,1.0275,2.6975,1.1575,2.8250,1.2850,2.9550,1.4125,3.0825,1.5425,0.0000,1.6700,0.1300,1.7975,0.2575,1.9275,0.3850,2.0550,0.5150,2.1825,0.6425,2.3125,0.7725,2.4400,0.9000,2.5675,1.0275,2.6975,1.1575,2.8250,1.2850,2.9550,1.4125,3.0825,1.5425,0.0000,1.6700,0.1300,1.7975,0.2575,1.9275,0.3850,2.0550,0.5150,2.1825,0.6425,2.3125,0.7725,2.4400,0.9000,2.5675,1.0275,2.6975,1.1575,2.8250,1.2850,2.9550,1.4125,3.0825,1.5425
PA:
0.0000,1.6700,0.1300,1.7975,0.2575,1.9275,0.3850,2.0550,0.5150,2.1825,0.6425,2.3125,0.7700,2.4400,0.9000,2.5675,1.0275,2.6975,1.1550,2.8250,1.2850,2.9525,1.4125,3.0825,1.5400,0.0000,1.6700,0.1300,1.7975,0.2575,1.9275,0.3850,2.0550,0.5150,2.1825,0.6425,2.3125,0.7700,2.4400,0.9000,2.5675,1.0275,2.6975,1.1550,2.8250,1.2850,2.9525,1.4125,3.0825,1.5400,0.0000,1.6700,0.1300,1.7975,0.2575,1.9275,0.3850,2.0550,0.5150,2.1825,0.6425,2.3125,0.7700,2.4400,0.9000,2.5675,1.0275,2.6975,1.1550,2.8250,1.2850,2.9525,1.4125,3.0825,1.5400
It took a while to notice but as you mentioned, there are small differences between AP and PA acquisition.
During writing this answer, my eyes dropped on an odd number of slices I have here and never had before  . I run into this answer from Jesper Andersson on JISCMail. He suggests not to delete any slices (as I used to before - because of multiband), but to use a different config file. So I have specified it within dwifslpreproc as `-topup_options " --config=b02b0_1.cnf " Although according to FSL web (topup/TopupUsersGuide - FslWiki), I don’t need to redirect to FSLDIR directory, it searches within automatically, It didn’t (it is just a minor advise for future readers). Answer from terminal when command run:
. I run into this answer from Jesper Andersson on JISCMail. He suggests not to delete any slices (as I used to before - because of multiband), but to use a different config file. So I have specified it within dwifslpreproc as `-topup_options " --config=b02b0_1.cnf " Although according to FSL web (topup/TopupUsersGuide - FslWiki), I don’t need to redirect to FSLDIR directory, it searches within automatically, It didn’t (it is just a minor advise for future readers). Answer from terminal when command run:
It looks like command was called with 2 config file arguments? From original 10 hours, time for Topup step increased to 91.6 hours 

 only to get the same warning and not producing files again.
only to get the same warning and not producing files again.
I also have one minor question. I couldn’t help but notice that the slspec file from -nocleanup (provided by dwifslpreproc) when using only .mif with SliceTiming info directly from .dcm looks different then mine from a script I made.  My matlab code (MBF stands for multi band factor, here it is 3). Data were measured as interleaved on Siemens Prisma 3T:
My matlab code (MBF stands for multi band factor, here it is 3). Data were measured as interleaved on Siemens Prisma 3T:
[~, slice_order] = sort(info.dicom_header.SliceTiming);
slice_order_reshape=reshape(slice_order,length(slice_order)/MBF,MBF);
slice_order_reshape=slice_order_reshape-1;
Thanks a lot for your time and any answers!
Stay happy and healthy,
Michaela.

 . I run into this answer from Jesper Andersson on
. I run into this answer from Jesper Andersson on 
 My matlab code (MBF stands for multi band factor, here it is 3). Data were measured as interleaved on Siemens Prisma 3T:
My matlab code (MBF stands for multi band factor, here it is 3). Data were measured as interleaved on Siemens Prisma 3T: