Dear MRtrix experts,
I’m experiencing some problems when running topup/eddy on denoised DWI data.
I have two DWI runs with reversed PE direction, each run has 30 directions with b=1000 and a few b=0 images.
This is what I have done:
1. denoise data
dwidenoise dwi.nii.gz dwi_denoised.nii.gz
dwidenoise dwi_revPE.nii.gz dwi_revPE_denoised.nii.gz
2. degibbs denoised data
mrdegibbs dwi_denoised.nii.gz dwi_denoised_unringed.nii.gz
mrdegibbs dwi_revPE_denoised.nii.gz dwi_revPE_denoised_unringed.nii.gz
3. create b0_revB0 image
fslroi dwi_denoised_unringed.nii.gz dwi_denoised_unringed_b0.nii.gz 0 1
fslroi dwi_revPE_denoised_unringed.nii.gz dwi_revPE_denoised_unringed_b0.nii.gz 0 1
fslmerge -t denoised_unringed_b0_revb0 dwi_denoised_unringed_b0.nii.gz dwi_revPE_denoised_unringed_b0.nii.gz
4. run topup
topup --imain=denoised_unringed_b0_revb0.nii.gz --datain=acqparams.txt --config=b02b0.cnf --out=denoised_unringed_b0_revb0_topup
5. create brain mask
applytopup --imain=dwi_denoised_unringed.nii.gz --inindex=1 --datain=acqparams.txt --topup=denoised_unringed_b0_revb0_topup --method=jac --out=dwi_denoised_unringed_cor
fslroi dwi_denoised_unringed_cor.nii.gz dwi_denoised_unringed_cor_b0 0 1
bet dwi_denoised_unringed_cor_b0.nii.gz dwi_denoised_unringed_cor_b0_brain -m -f 0.2
6. run eddy with topup
eddy --imain=dwi_denoised_unringed.nii.gz --mask=dwi_denoised_unringed_cor_b0_brain_mask.nii.gz --index=index.txt --acqp=acqparams.txt --bvecs=dwi.bvec --bvals=dwi.bval --topup=denoised_unringed_b0_revb0_topup --out=dwi_denoised_unringed_eddy
This is what the input DWI (without any preprocessing) looks like (dwi.nii.gz):
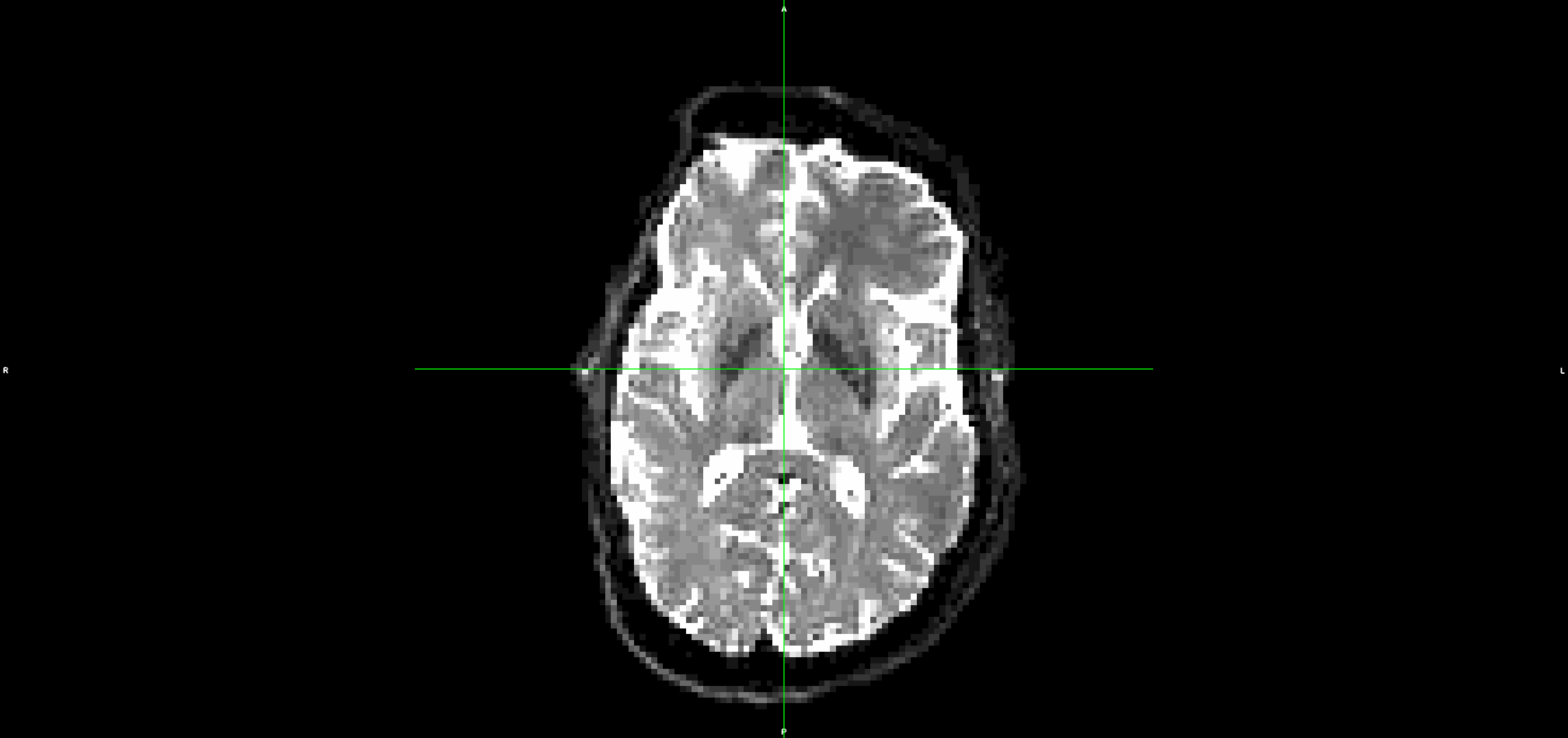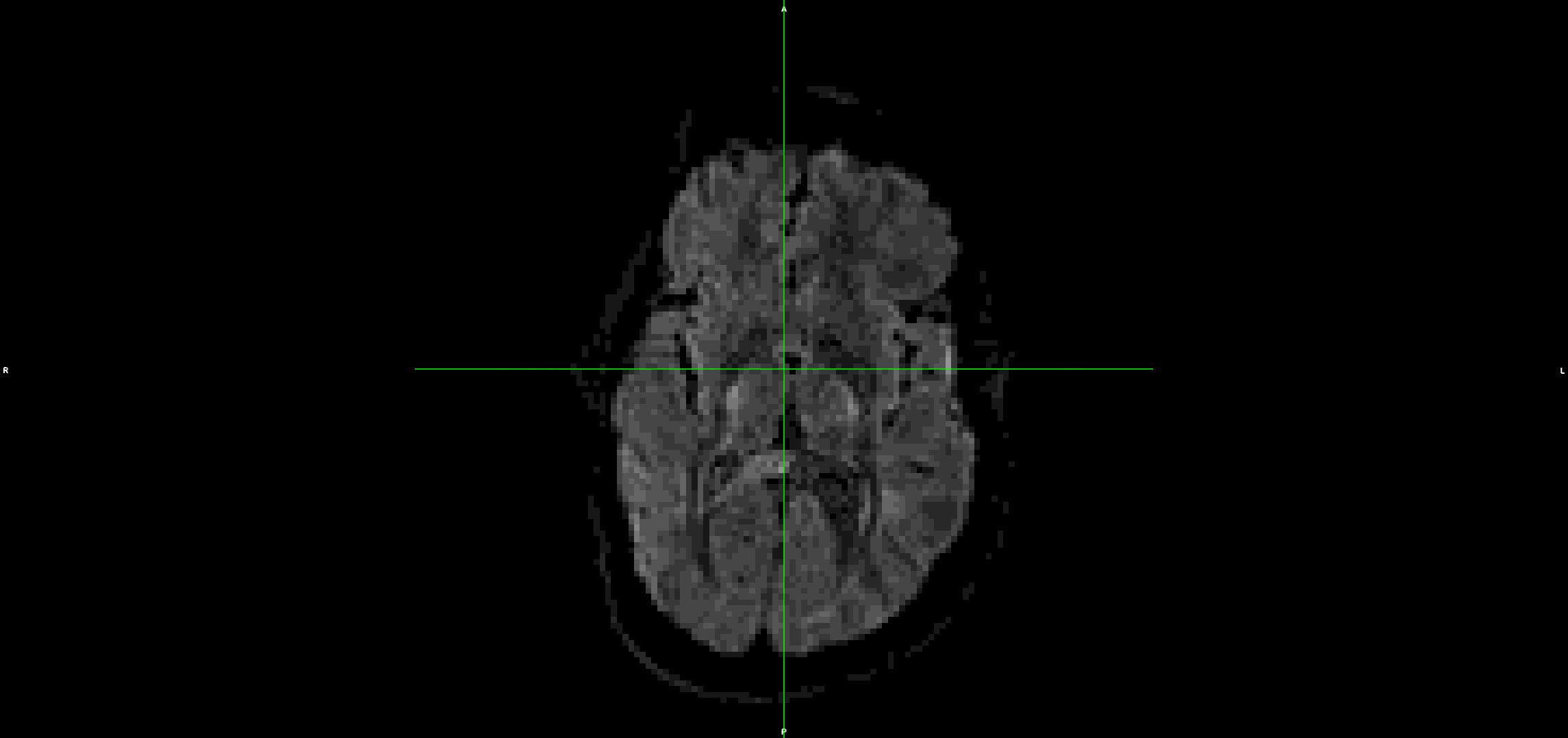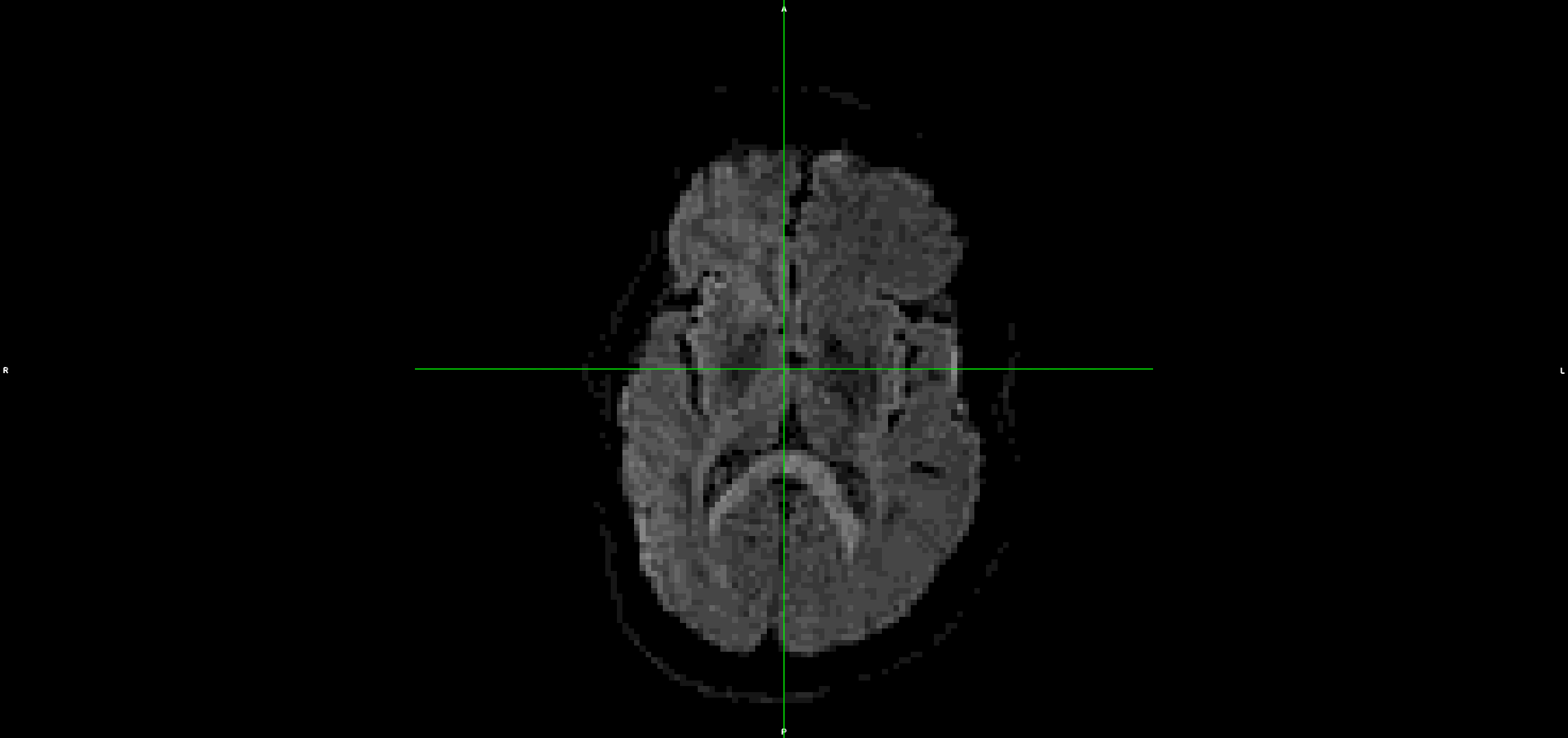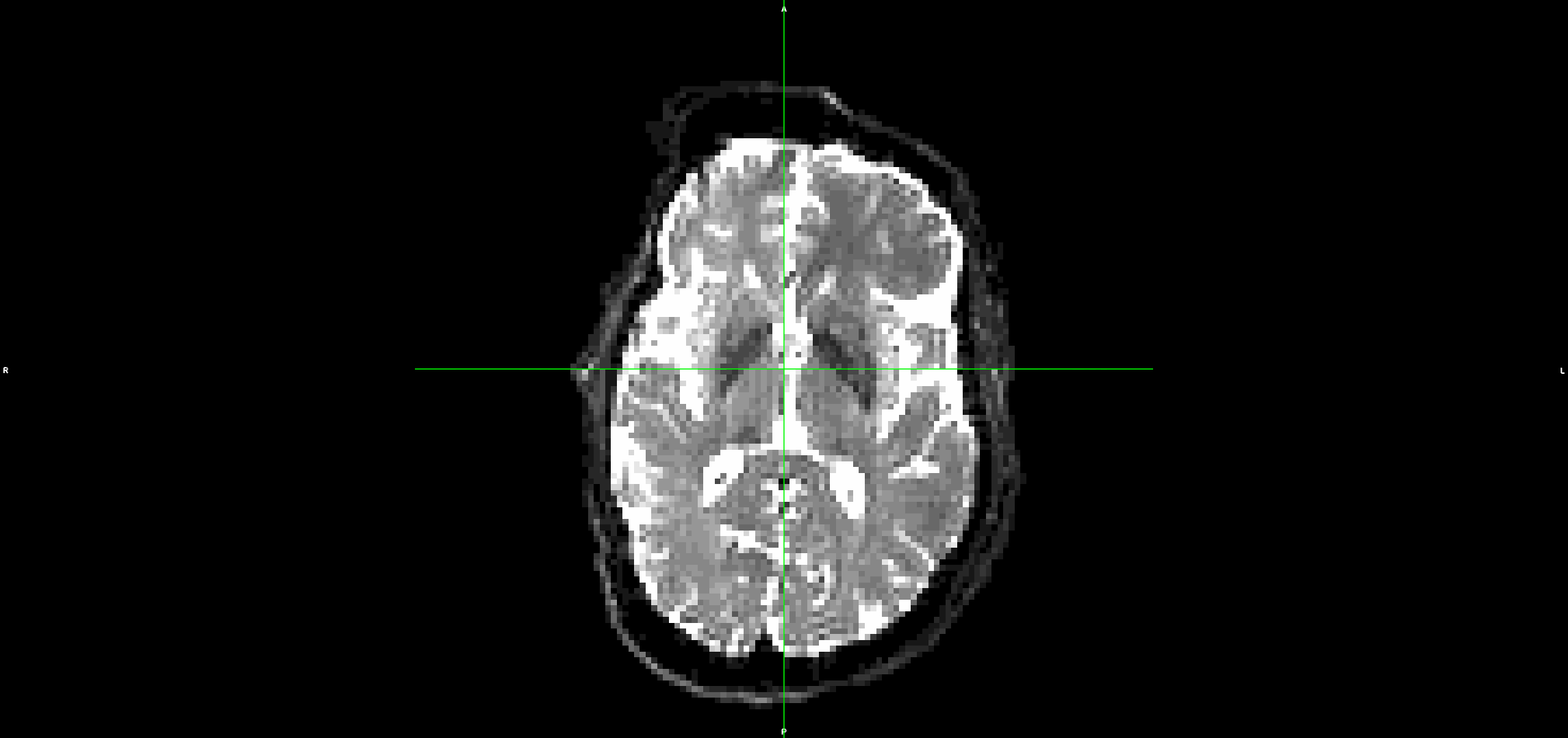
This is the output from topup/eddy (dwi_denoised_unringed_eddy.nii.gz):
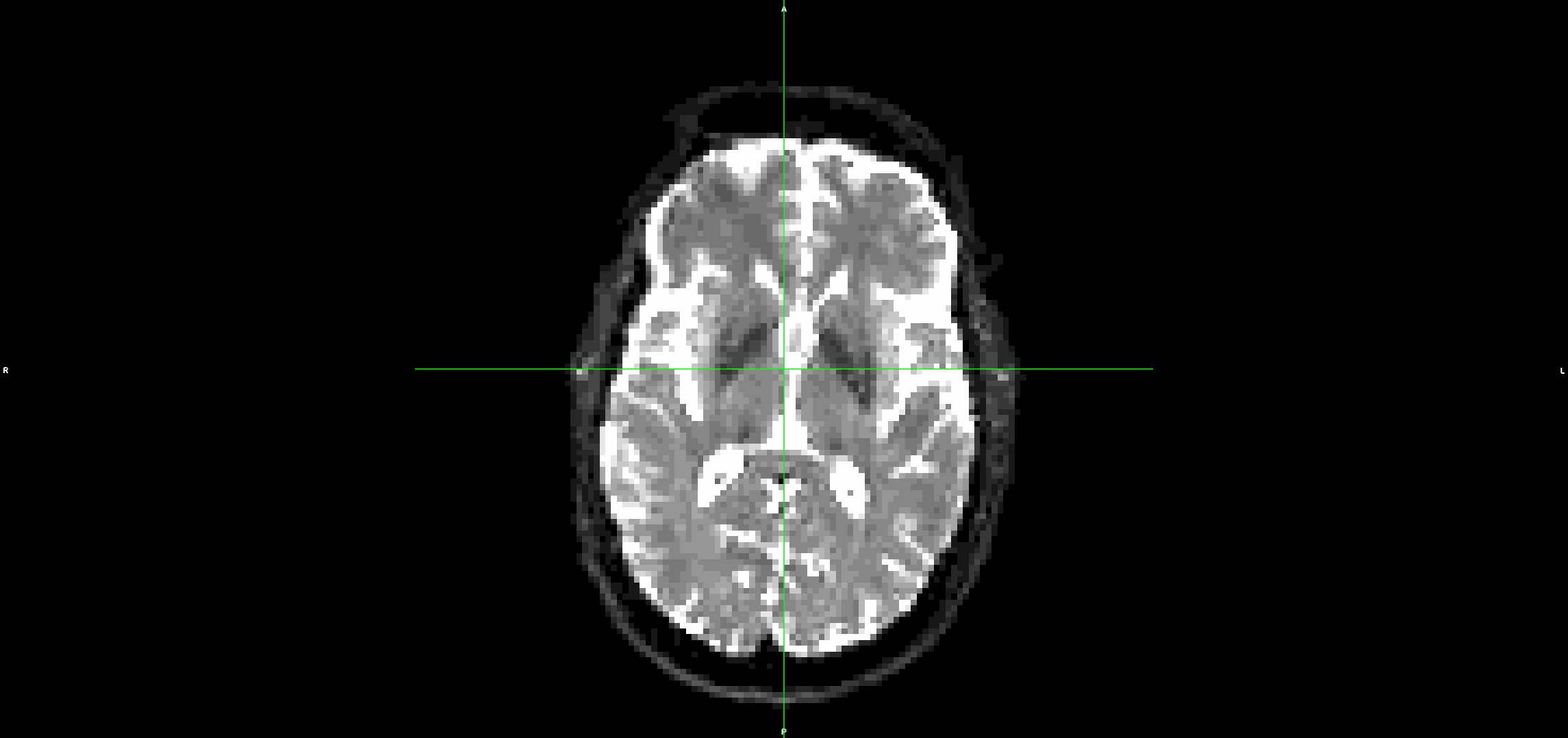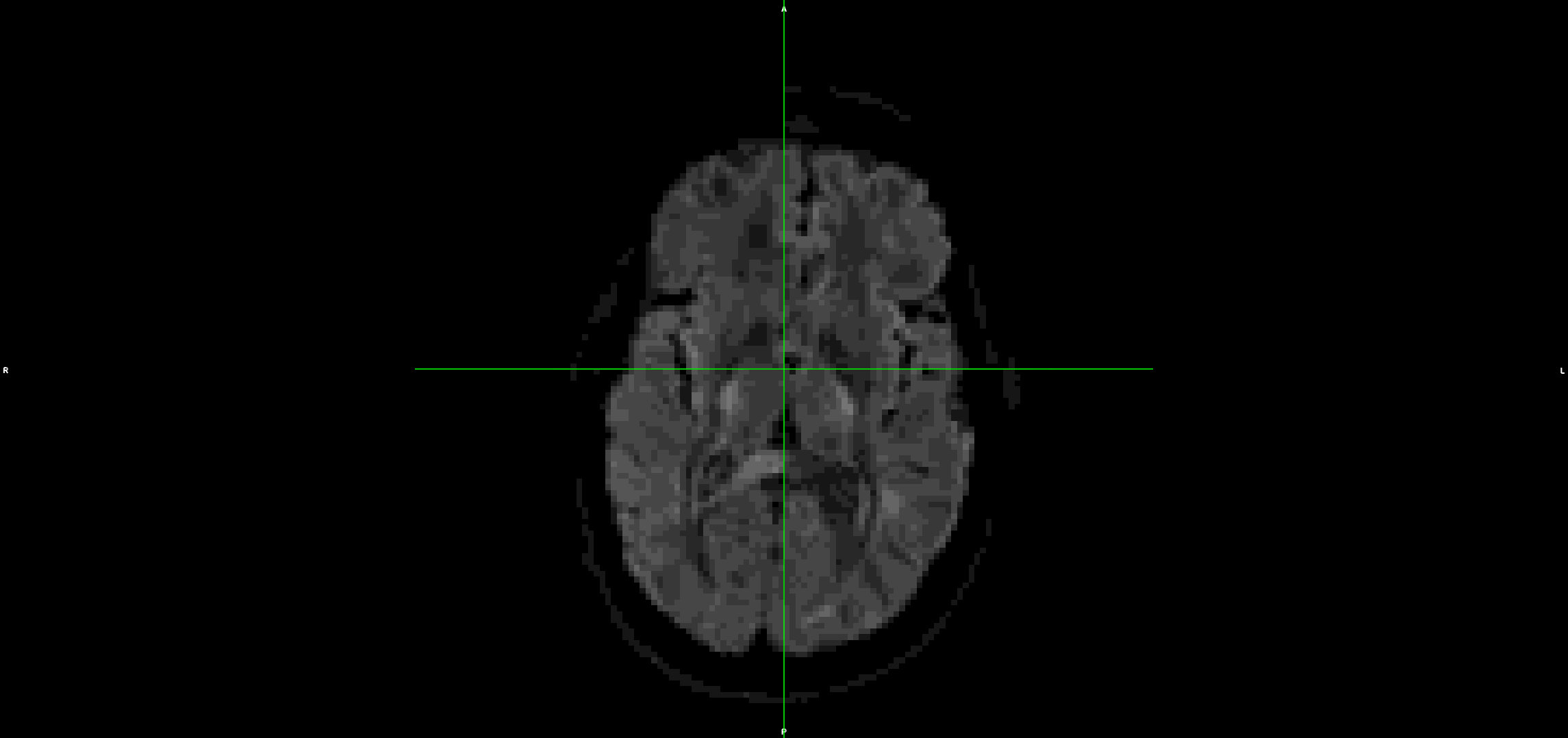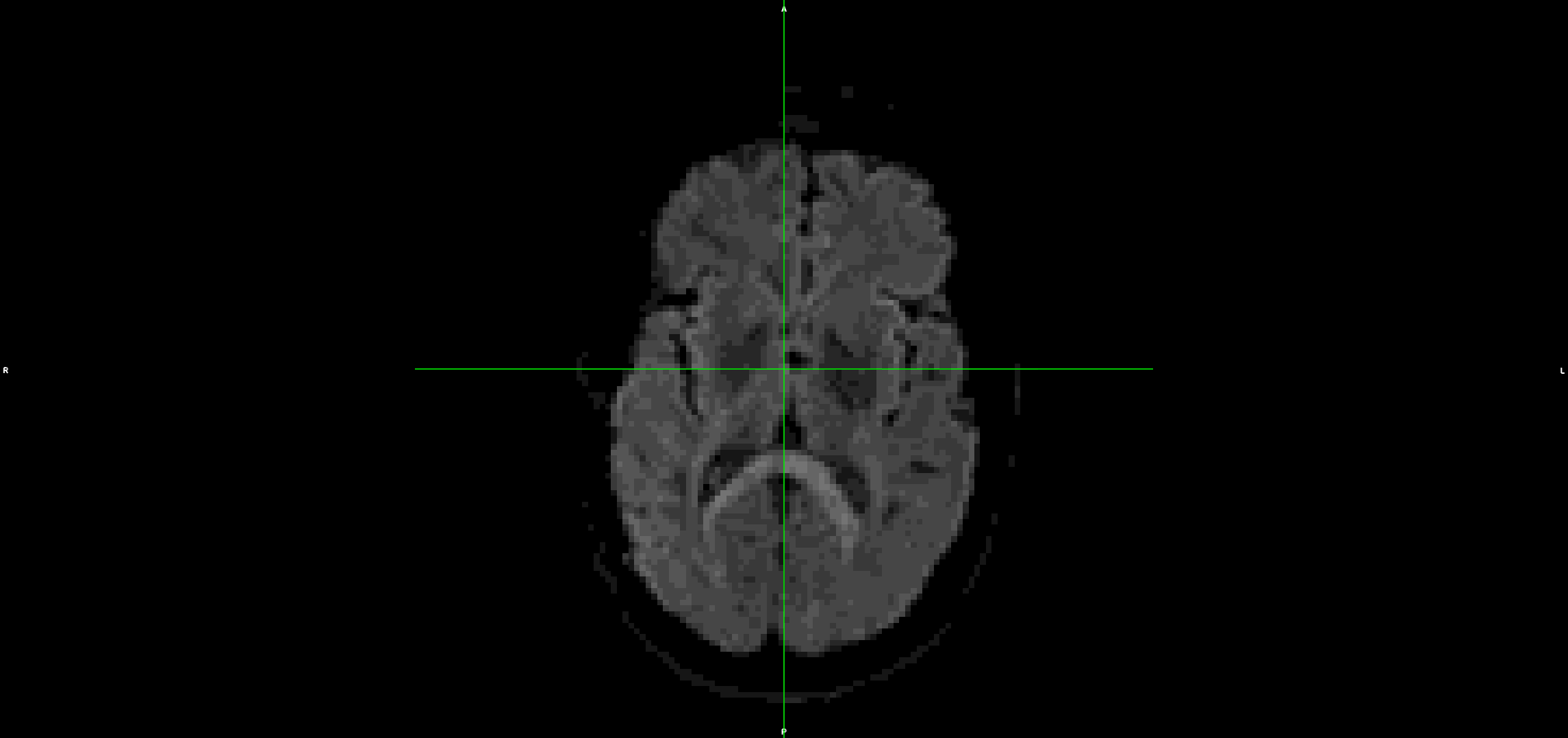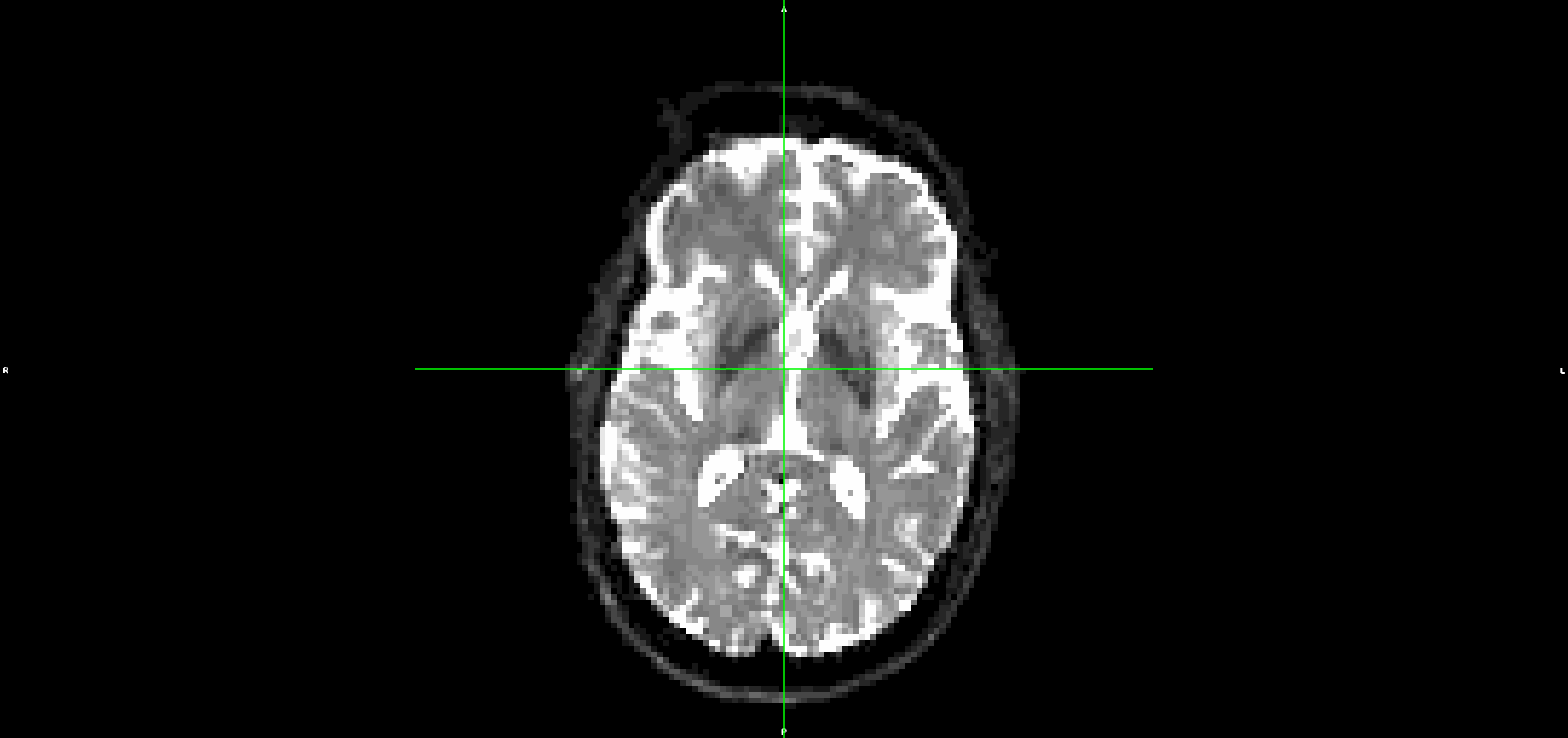
It looks to me that the movement artefacts after running topup+eddy are similar to the raw data.
If I run the same steps, but without denoising and unriniging the data (so starting from step 3 above), the eddy output looks much better with less residual movement:
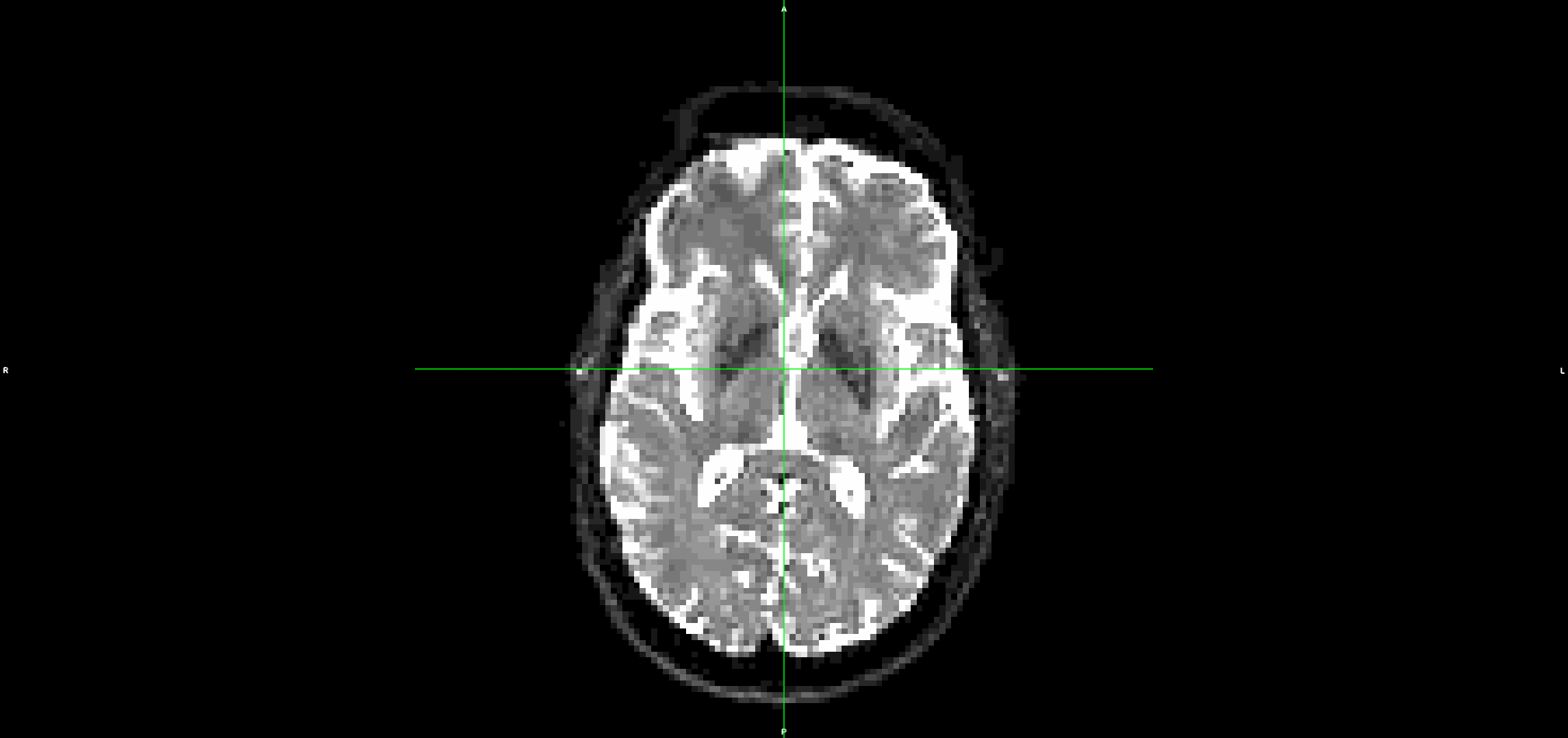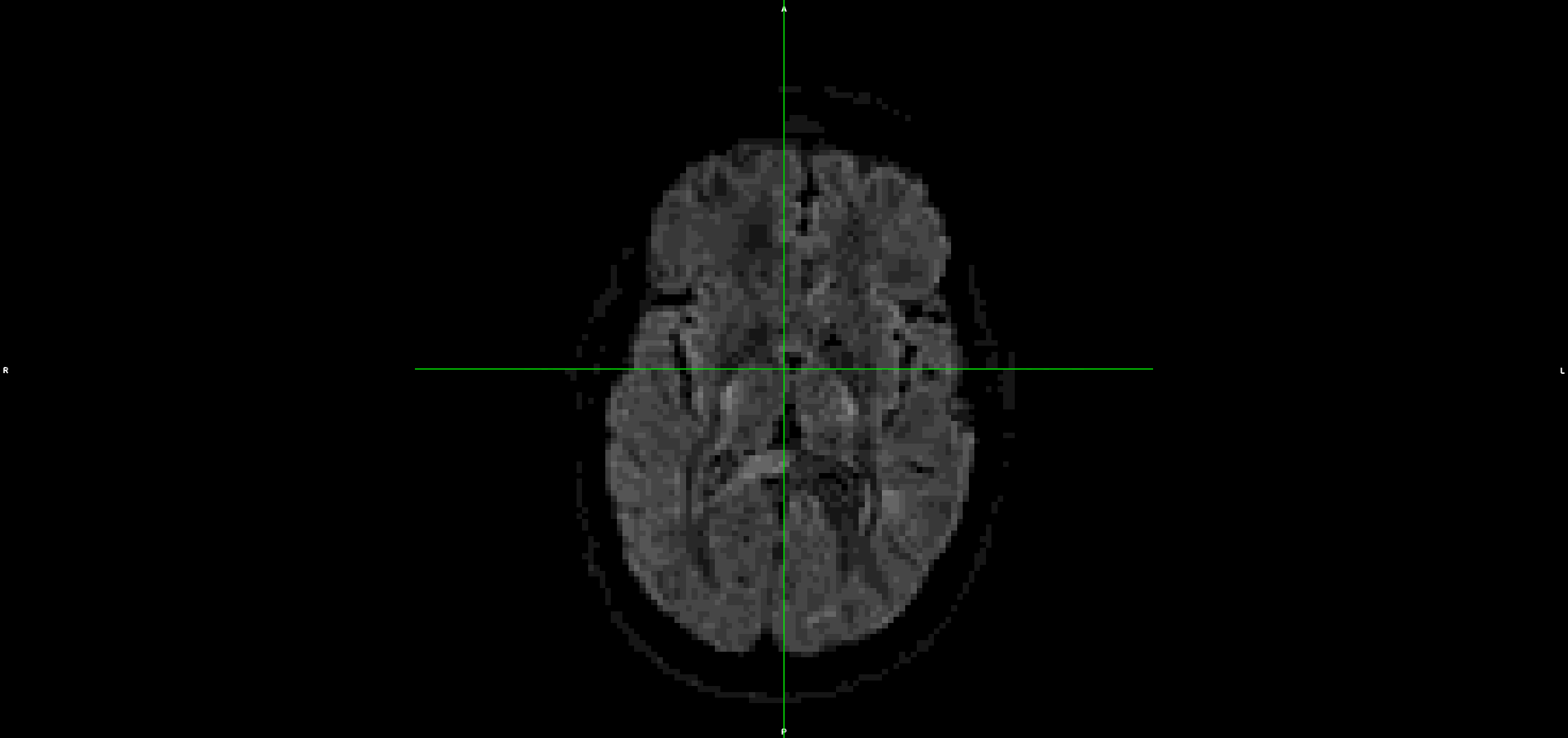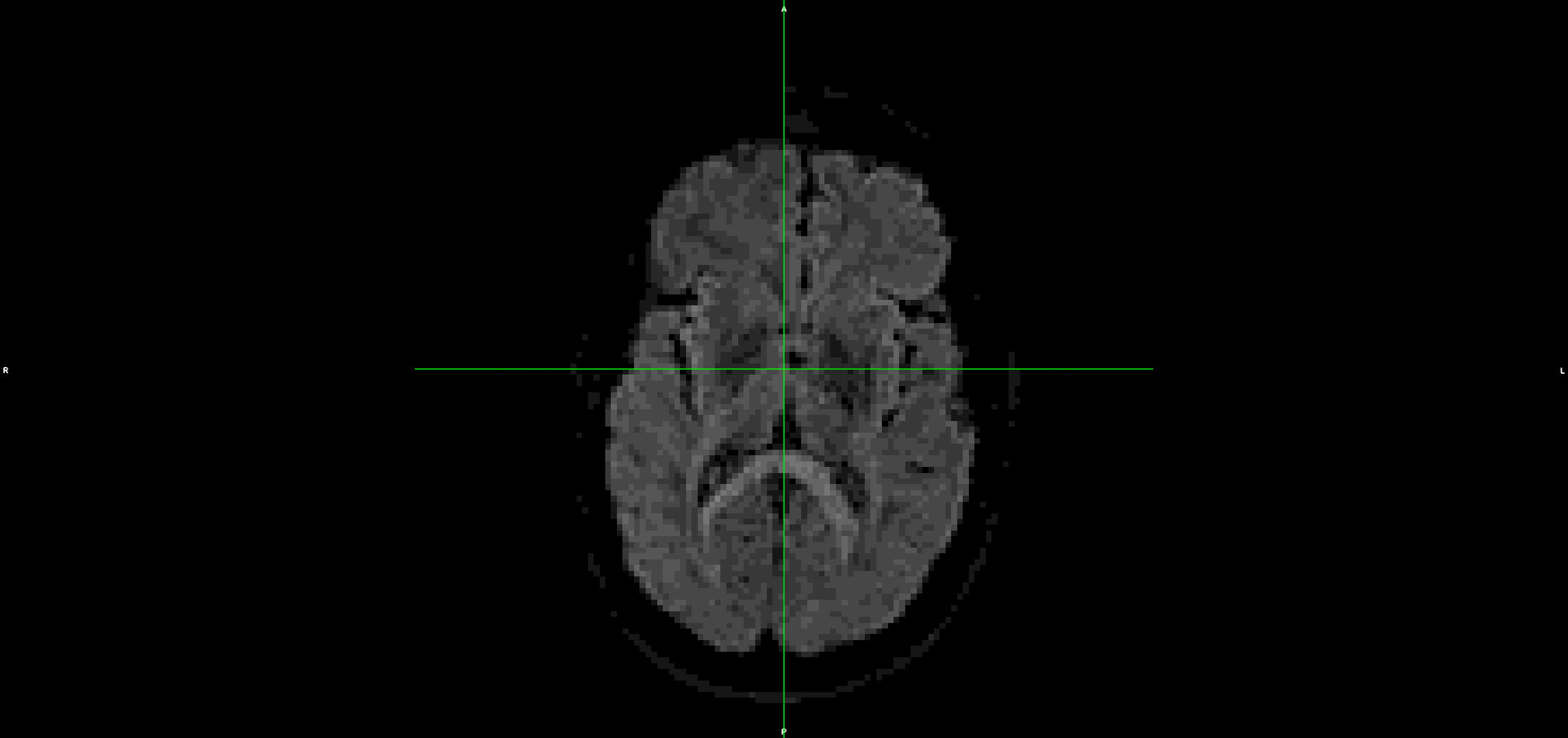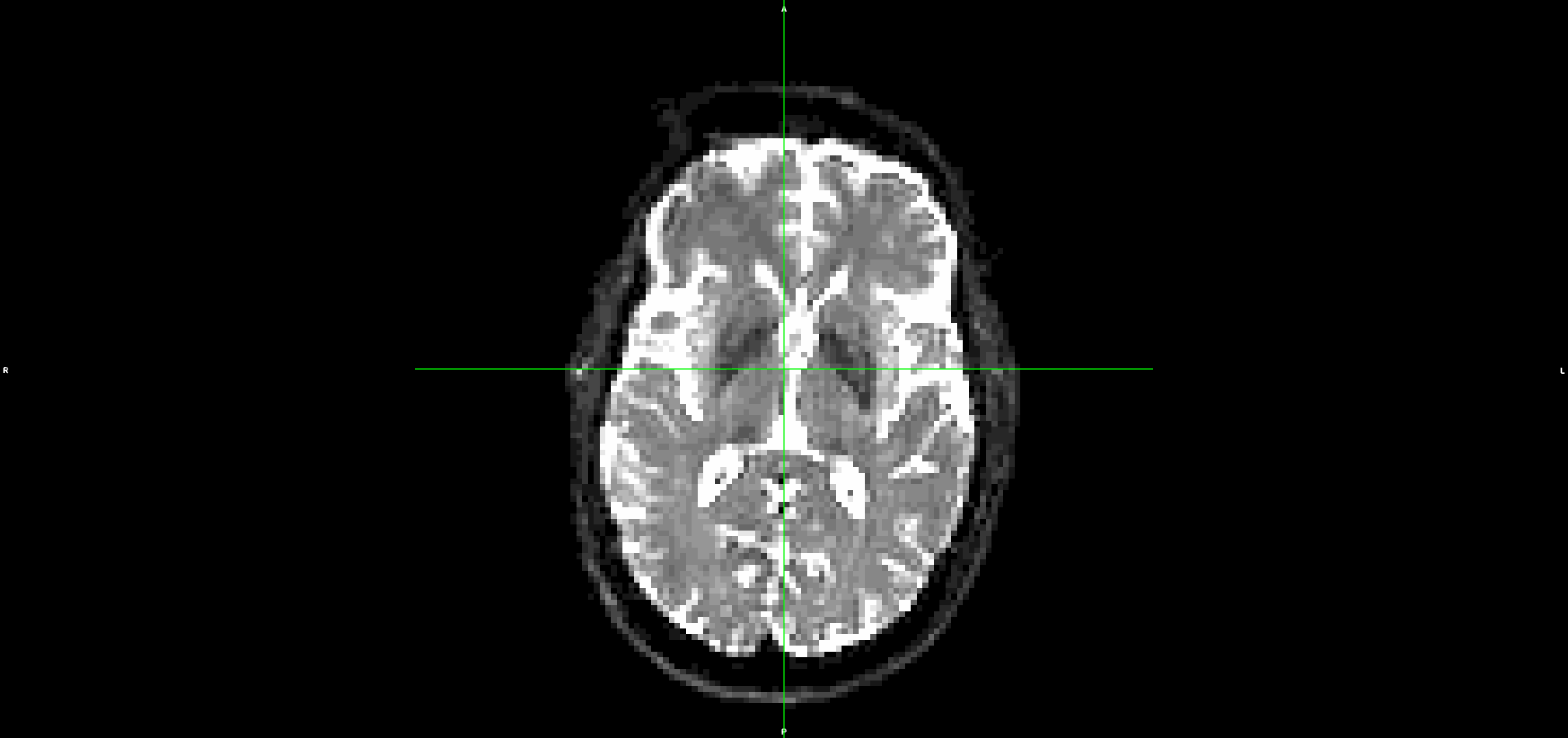
I have also tried it without topup and the problem is the same.
Has anyone experienced a similar issue? Is it possible that applying eddy on denoised data leads to something weird for some datasets?
Thanks a lot in advance for your help!
Kind regards,
Julia