Hi all! Newbie MD student here from Sydney.
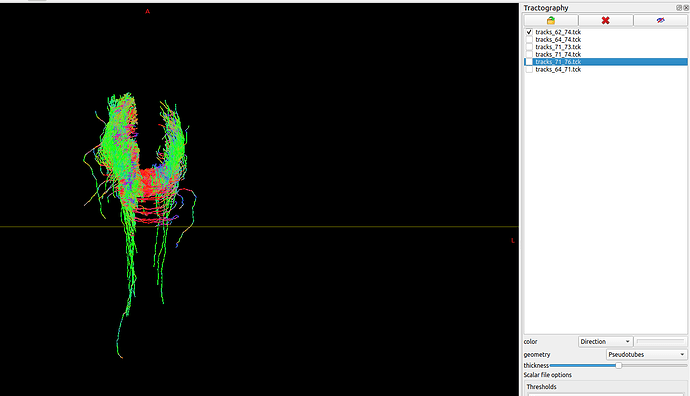
I’m exploring the nodes of the default mode network, and wanted to make a visualisation of the streamlines of particular node pairs, taking a unilateral view.
I’m confused since although most of my tracts appear appropriate (besides the expected spurious fibres), the Rostral Anterior Cingulate Gyrus to Medial-Orbital Frontal Gyrus nodes on the R side (i.e. DKA atlas nodes 62 and 74) has significant tracking to the L side.
Is this expected behaviour? Looking at my parcels.mif, the regions are as expected. It seems strange that so many streamlines went across the corpus callosum to the other side?
Tractography command: tckgen -act ../5ttgen/5tt_coreg.mif -backtrack -seed_gmwmi ../5ttgen/gmwmSeed_coreg.mif -nthreads 8 -maxlength 250 -cutoff 0.06 -select 10000000 ../fod_wm.mif tracks_10M.tck
Connectome: tck2connectome -symmetric -zero_diagonal -scale_invnodevol -assignment_all_voxels ../tractography/tracks_10M.tck parcels.mif parcels.csv -out_assignment assignments_parcels.txt
Node-Node tractography: connectome2tck tracks_10M.tck ../connectome/assignments.txt tracks_62_74.tck -nodes 62,74 -exclusive -files single