Dear experts,
I am performing tractography with mrtrix3 v3.0.1 and using a custom parcellation to get a connectome using tck2connectome. When I check my results using connectome2tck with streamline assignments from tck2connectome, i get noisy fibers in other parts of the brain (that don’t start or terminate in either node I’m looking at). I haven’t reproduced this error yet (by repeating the tck2connectome and connectome2tck runs) but I was hoping to see if anyone else has had this error because it seems kinda serious. I’ve followed the multi-shell pipeline (I’m using multi-shell data) with denoise, topup, eddy, bias correction and finally generating 25 million streamlines using tckgen.
these are the commands I’m running:
tckgen wm.mif 25M.tck -act 5TT.nii -backtrack -crop_at_gmwmi -seed_dynamic wm.mif -minlength 10 -maxlength 250 -select 25M -mask brainmask.nii -output_seeds succeeds.txt -force -nthreads 8
tcksift2 ${numfibers}.tck wm.mif sift_weightfactor.txt -act r5TT.nii -fd_scale_gm -force -nthreads 12
tck2connectome 25M.tck parc.nii parc_streamweights.csv -tck_weights_in sift_weightfactor.txt -assignment_radial_search 2 -zero_diagonal -out_assignments streamlineassignment.txt -force -nthreads 12
connectome2tck 25M.tck streamlineassignment.txt subj -nthreads 12
This is the error that I’m getting:
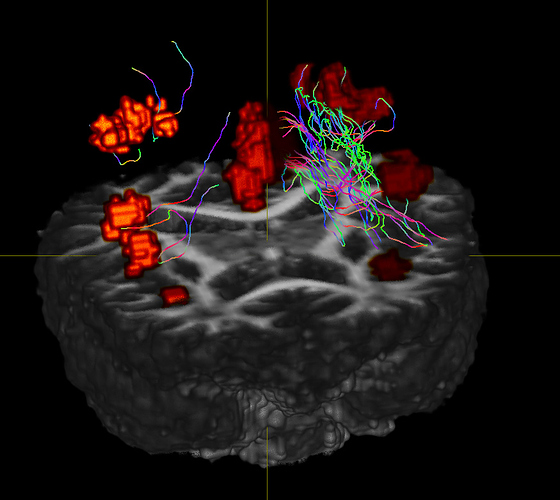
As you can see, I get the streamlines from the nodes at the front left (right on image) but I also get tracts from the back of the brain as well that don’t seem to come from anywhere.
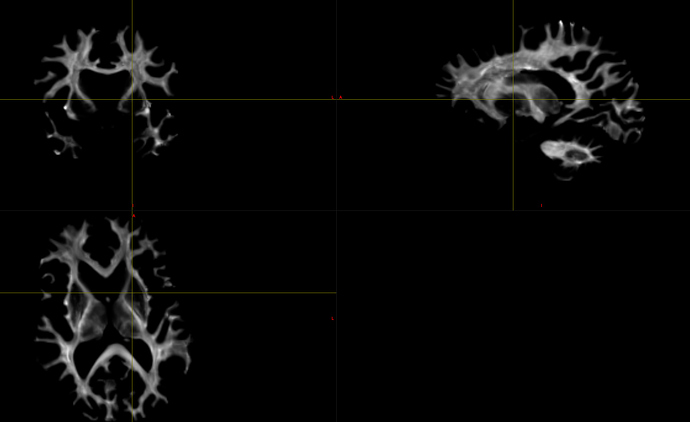
There’s nothing wrong with the tractography (that I can tell):
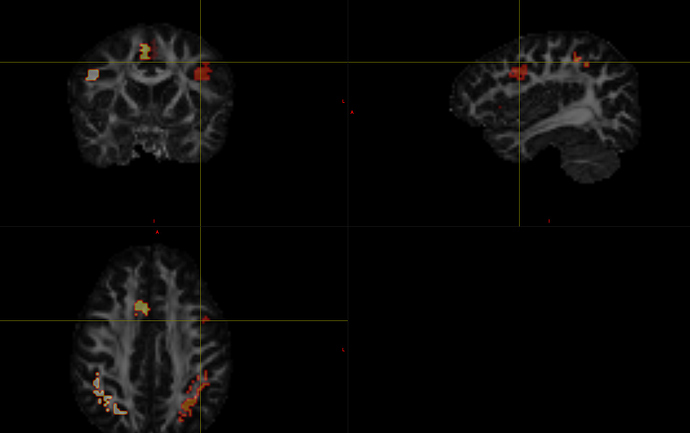
And here’s my parcellation:
The nodes are custom masks informed from fmri. They are discontiguous because parts of the nodes are in WM (so I cut those out) but I don’t see why this should be causing an issue.
This error is common among subjects as well.
I would appreciate some advice here on how to proceed; whether I just use thresholding to get rid of the noisy fibers, or what the cause of the error might be (if it is indeed an error?)
Best,
Nikitas Koussis