Greetings all,
I’m hoping you can help me out here. I’m working on a dataset of ex-vivo mouse brains. We collected 0.2mm isotropic imaging with 3 shells (b1000, 2000, 3000 +b0) and 64 directions per shell. Initial preprocessing (including reorientation of the images and eddy correction) was done using TORTOISE.
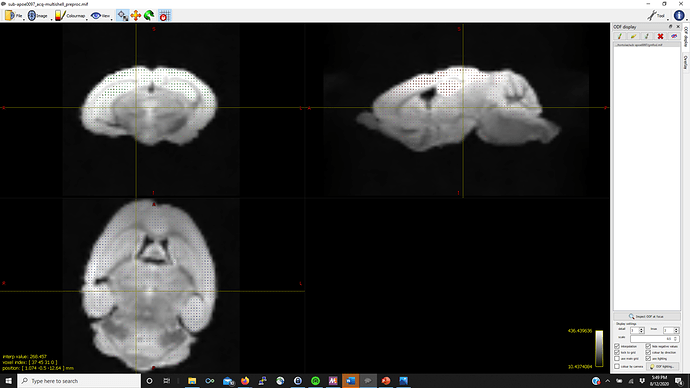
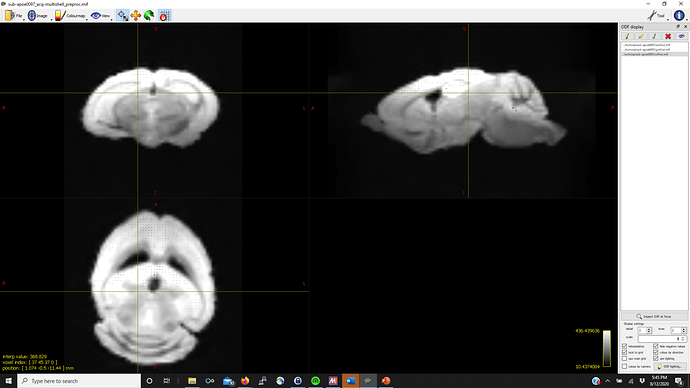
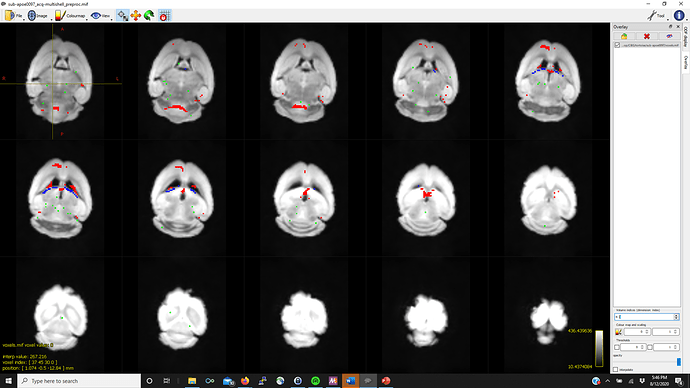
I’m trying to run the FBA pipeline described in the documentation. dwi2response dhollander produces reasonable WM FODs but struggles to detect good representative CSF voxels. As such, in several of the mice, mtnormalise has a negative tissue balance such as
[WARNING] For input "sub-apoe0097" (returncode = 1):
mtnormalise: [WARNING] existing output files will be overwritten
mtnormalise: performing log-domain intensity normalisation... []
mtnormalise: [ERROR] Non-positive tissue balance factor was computed. Balance factors: 4.3531205638674342 3.559592222662189 -0.41592815237490682
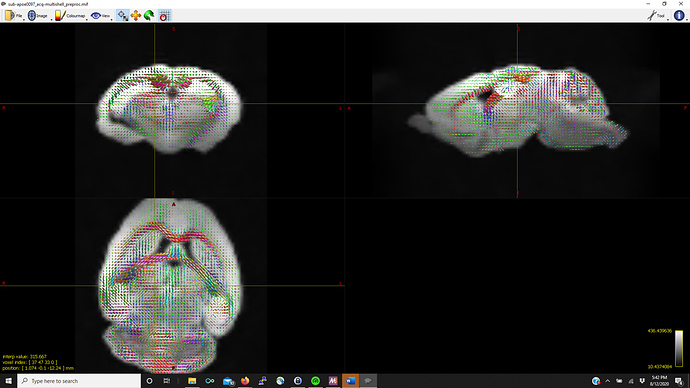
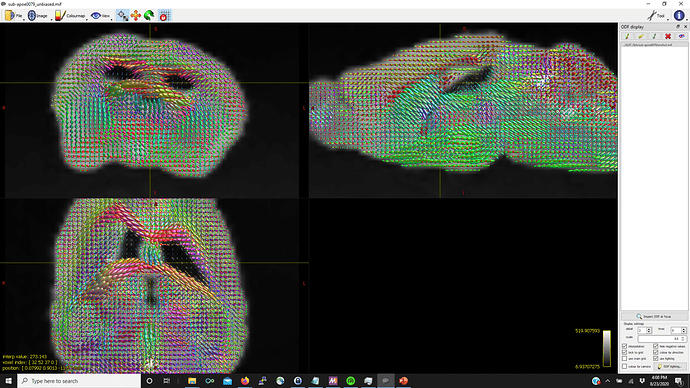
However, the gray matter FODs are fairly reasonable and well-placed (in contrast to other posts on the community) in almost all of the mice.
I know there was a suggestion in a post somewhere (not finding it right now) to use a 2 tissue WM-CSF mtnormalise but I’m wondering if a WM-GM mtnormalise would be a valid strategy.
Thanks in advance (pictures below for WM, GM, CSF FODs and the voxel image from dwi2response from a representative mouse that fails mtnormalise)