Dear Experts,
I have conducted FBA on my subject data and extracted FA images using MRtrix. After aligning the FA images to MNI space, I performed SPM’s TFCE analysis in MATLAB.
I would like to ask: if I want to compare the results of FBA with the results from SPM, would it be better to align the subject data to MNI space before running FBA, or should I align the wmfod_template.mif to MNI space instead?
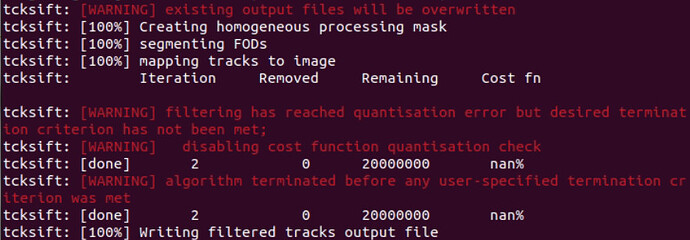
Currently, I have adopted the second approach: aligning the wmfod_template to MNI space. I then followed the official tutorial to complete the subsequent FBA steps. However, I got stuck at Step 19, where I couldn’t proceed with tcksift. Could you please advise me on which part of my steps needs adjustment?
And these are the commands I’ve done to aligning wmfod_template to MNI space:
fslmaths ../wmfod_template_l0.nii.gz -nan l0image.nii.gz
flirt -ref ../../../JHU/MNI152_T1_1mm_brain.nii.gz -in l0image.nii.gz -out l02MNI.nii.gz -omat l02mni.mat
fnirt --ref=../../../JHU/MNI152_T1_1mm_brain.nii.gz --in=l0image.nii.gz --aff=l02mni.mat --cout=warps_l02mni
ls
warpinit l0image.nii.gz identity_warp_wmfod_template.nii
applywarp --ref=../../../JHU/MNI152_T1_1mm_brain.nii.gz --in=identity_warp_wmfod_template.nii --warp=warps_l02mni.nii.gz --out=mrtrix_warp_template2mni.nii.gz
ls
mrconvert identity_warp_wmfod_template.nii -coord 3 0 - | mrcalc - 0 -mult 1 -add identity_warp_wmfod_template_mask.nii
applywarp --ref=../../../JHU/MNI152_T1_1mm_brain.nii.gz--in=identity_warp_wmfod_template_mask.nii--warp=warps_l02mni.nii.gz --out=mrtrix_warp_template2mni_mask.nii.gz
applywarp --ref=../../../JHU/MNI152_T1_1mm_brain.nii.gz --in=identity_warp_wmfod_template_mask.nii --warp=warps_l02mni.nii.gz --out=mrtrix_warp_template2mni_mask.nii.gz
ls
mrcalc mrtrix_warp_template2mni_mask.nii.gz 1 nan -if mrtrix_warp_template2mni.nii.gz -mult mrtrix_warp_template2mni_valid.nii.gz
mrtransform ../wmfod_template.nii.gz -warp mrtrix_warp_template2mni_valid.nii.gz -template ../../../JHU/MNI152_T1_1mm_brain.nii.gz wmfod_template_MNI.nii.gz -reorient_fod yes
mrinfo wmfod_template_MNI.nii.gz
Here is the error in tcksift:
Best,
Vivian