Hi guys,
Since dwifslpreproc calls FSL’s eddy to do its thing, I figured I’d ask here about it here as well, to get more perspectives on the issue.
I have been performing eddy on some scans I have. The subjects are animal brains scanned ex-vivo (therefore very little movement between volumes/directions) so I expect little changes after eddy (my supervisor told me that we don’t need to apply topup for these scans, but nonetheless we still have to apply eddy before FDT and getting DTI values for stats). So I performed eddy using only the mandatory inputs (bvals, bvecs, acqp, index, mask and dti data) and for most scans this worked fine. I haven’t used any additional option like repol or anything. Only the mandatory commands.
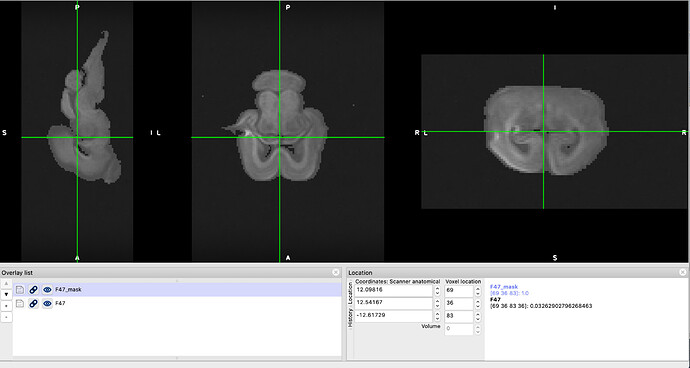
The masks were not prepared using bet, though. Since bet was giving me aberrant results (apparently bet works well for human brains on skulls, and I’m working with animal scans without skull, namely ferrets, sheep and mice), I opted for using select_dwi_vols to extract all volumes for b4000, average them using fslmaths and then thresholding using fslmaths -thr at a value that created a mask capturing the whole brain. As mentioned before, this apparently worked fine for most scans as I could see in their eddy_out files.
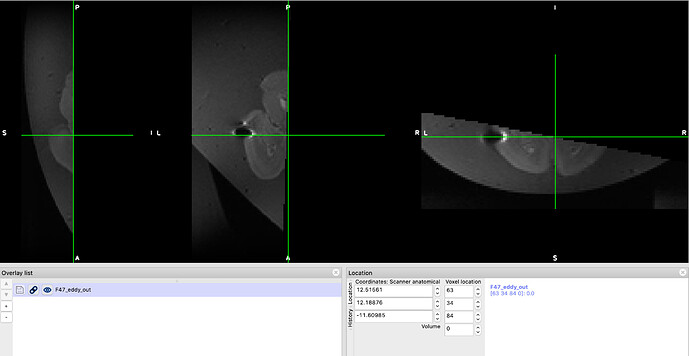
However on some scans I experienced loss of a substantial amount of data, even when the subject showed little to no movement in the acquisition. The only movement experienced during the acquisition I show here, for example, is a slow shift of the brain towards the left, on a total of 81 volumes, which the mask is a bit bigger towards the sides (see top image below). But the output is missing half the data (see bottom image below). How can this be prevented?
On other scans I lost a bit of data even when the subject experienced no movement during acquisition (brain embedded in agar). Could in this particular case be due to field inhomogeneities (that should have been corrected using topup even when my supervisor told me we could safely skip it)? How sensitive is eddy to these inhomogeneities? I’m asking because the loss of data in that case coincided with a region that is a bit dimmer than the rest of the scan. But to be honest that’s just an observation since I’m really not sure what is causing the issue, and I find it strage because in other cases I experienced no loss of data even when there were these differences in brightness in some regions. The acquisition protocol is exactly the same for animals from the same species, but the process has failed in only a handful. The eddy command is also the same for all of them (since I performed eddy in a HPC using a script with a loop).
module load fsl
eddyfolders=`find $1 -name '*' -type d -maxdepth 1 -mindepth 1`
for SPEC in $eddyfolders
do
cd "$SPEC"
eddy --imain="${SPEC}.nii" \
--mask="${SPEC}_mask.nii" \
--index="${SPEC}_index.txt" \
--acqp="${SPEC}_acqp.txt" \
--bvecs="${SPEC}.bvec" \
--bvals="${SPEC}.bval" \
--out=eddy_out \
--verbose
cd ..
done
Thanks for your help and insights!!