Dear MRtrix experts,
I am processing some HCP data and I am interested in the differences between probabilistic and deterministic whole brain tractograms generated using 3M streamlines.
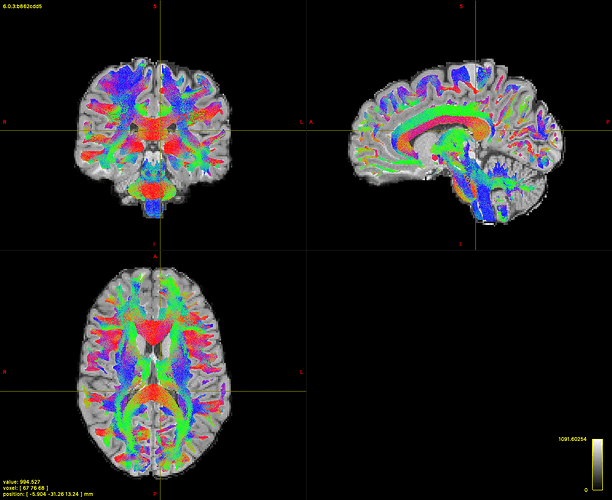
As probabilistic algorithm I used the iFOD2 and I mainly followed the pipeline available here. Everything looks fine and I am happy with that. Also when I filter the “non-connecting” streamlines (i.e. those that are not connecting 2 valid grey matter regions according to the standard Desikan-Killiany parcellation in 84 parcels plus the brainstem) I do not loose many streamlines: I go from 3M to 2749855 which is reasonable and all the white matter is covered (see picture).
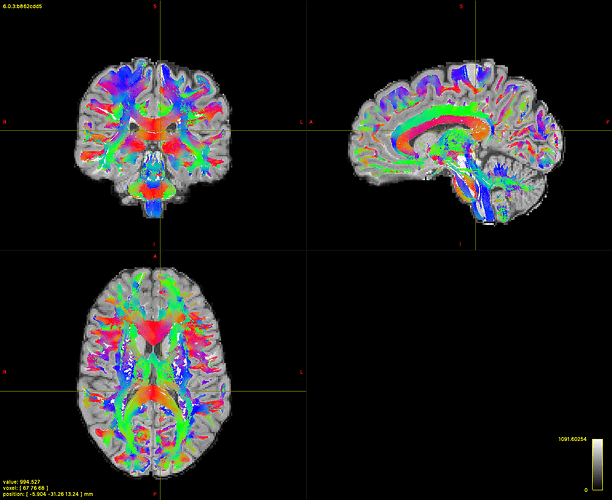
Something different happens when I use SD_stream… I tried both with and without ACT, seeding in the white matter mask or only at the grey-white matter boundary, changing angles, step size and cutoff values, but the best I was able to obtain starting from the same FOD used for iFOD2 is this
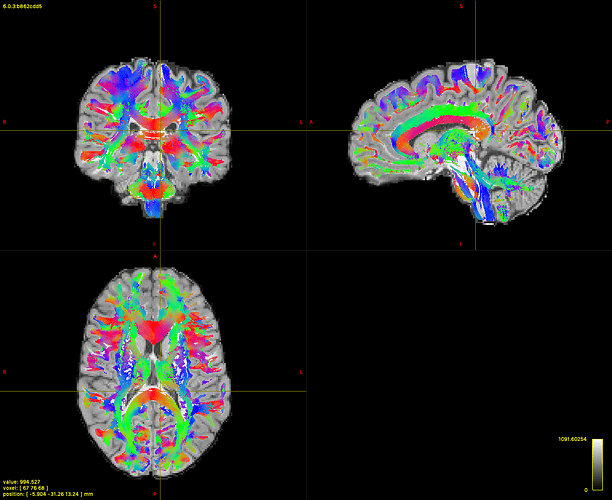
Which when I filter the non-connecting streamlines reduces to this
I do not loose a lot of streamlines because I end up with 2814529, but as you can see a large portion of the splenium of the corpus callous is not covered as well as many voxels in the brainstem and in areas where we have a lot of crossing.
I am aware of the limitations of deterministic tracking, but is there a way to obtain something better? Are there any parameters values that you know of that can help me improve my results? I suppose there is a way but I might not understand the algorithm enough to find the best choice. 
As reference here are the command lines I’ve used to generate the tractograms in the images and filter out the non connecting streamlines based on the parcellation.
tckgen FOD/WM_FODs_norm.nii.gz iFOD2_ACT_3M_hcp.tck -act 5TT/5TT.nii.gz -backtrack -crop_at_gmwmi -seed_gmwmi 5TT/5TT_mask.nii.gz -select 3M -maxlength 250 -minlength 5 -cutoff 0.06
tck2connectome Tractography/iFOD2_ACT_3M_hcp.tck -out_assignments Connectome/3M_iFOD2_ACT_assignment.txt Connectome/nodes_fixSGM.nii.gz Connectome/iFOD2_ACT_3M_connectome.csv -symmetric
connectome2tck -force Tractography/iFOD2_ACT_3M_hcp.tck Connectome/3M_iFOD2_ACT_assignment.txt Tractography/iFOD2_ACT_3M_hcp_connecting.tck -files single -keep_self -nodes 1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85 -exclusive
tckgen -algorithm SD_Stream FOD/WM_FODs_norm.nii.gz SD_STREAM_ACT_3M.tck -act 5TT/5TT.nii.gz -crop_at_gmwmi -seed_gmwmi 5TT/5TT_mask.nii.gz -select 3M -maxlength 250 -minlength 5 -cutoff 0.06 -angle 60 -step 0.25
tck2connectome Tractography/SD_STREAM_ACT_3M.tck -out_assignments Connectome/3M_SD_STREAM_ACT_assignment.txt Connectome/nodes_fixSGM.nii.gz Connectome/SD_STREAM_ACT_3M_connectome.csv -symmetric
connectome2tck -force Tractography/SD_STREAM_ACT_3M.tck Connectome/3M_SD_STREAM_ACT_assignment.txt Tractography/SD_STREAM_ACT_3M_connecting.tck -files single -keep_self -nodes 1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85 -exclusive
Thank you,
Simona
 ) Regardless, I’ve noticed this and similar problems with SD_STREAM too; it’s not suitable for most tractography jobs I’m afraid. If you’re looking for something “in between”, you can play around with the
) Regardless, I’ve noticed this and similar problems with SD_STREAM too; it’s not suitable for most tractography jobs I’m afraid. If you’re looking for something “in between”, you can play around with the