Hi all,
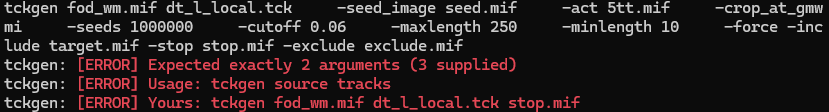
I want to track the dentatothalamic tract (DRTT) using the tckgen command. The seed is the dentate. The target is the contralateral thalamus. Now, I wish the fibertracking stops once it enters into the contralateral thalamus. So I copy the “target.mif” file and rename it “stop.mif”. Then, I used the following code, but it returned an error as shown in the screenshot. But if I deleted the “-stop stop.mif”, it could run normally. Could you help me find a solution? Thanks!!!
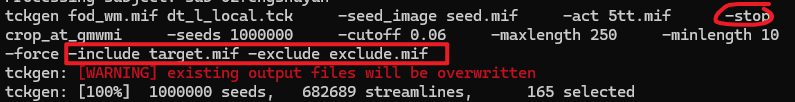
I find if I just type in “-stop” without a following ROI file, it can run normally. Does this mean that the “stop” option cannot be followed by an ROI file?
Hi Bin,
That’s correct, the -stop option will not accept an input image, rather, it flags to stop propagating a streamline once it has traversed all include regions.
If the thalamus region is labelled as SGM in the 5tt.mif image, then streamlines should terminate within the thalamus. As such, the stop option might be redundant.
Does the dt_l_local.tck output look sensible?
Cheers,
Arkiev
PS: This paper might be of interest for DRTT reconstruction…