Dear Mrtrix Experts,
I have an ex-vivo human spinal cord dataset. They are with 2 bva0 and 750, and 1500, each 12 directions. I’m trying to get a good response function in order to generate CSD map and Tractography. I would like to use Mrtrix 3 to do so.
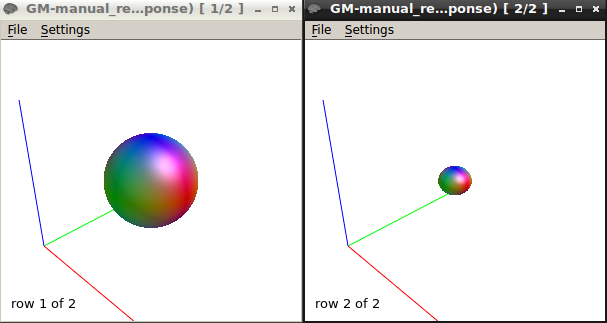
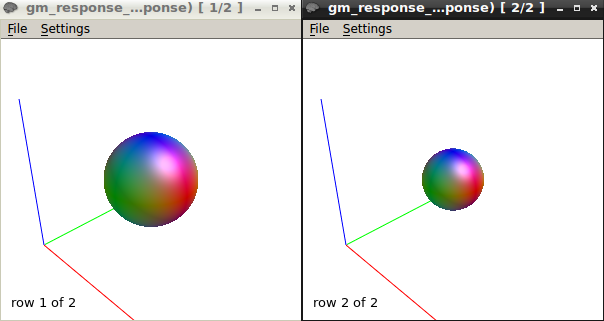
I’ve used the dwi2response tournier , dhollander, fa and manual algorithm using the whole cord or only selecting for the single fibre orientation WM tract areas. I am still not satisfied about the outcome of response function. I think I should aim to get a response function close to a flat oblate shape in order to get a good FODs? Could you recommend if this can be better performed?
I have done the following steps:
A. response calculation using the whole cord mask
-
I converted the dicom to nifti format using (MIPAV) for first sample 3 shells b0,750 &1500. For second sample (b0&3000) I used mrconvert dwi2nifti
-
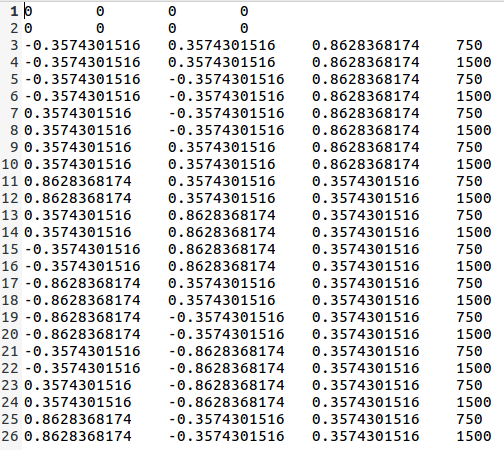
For the gradient table, I swapped the gradient from xyz to zyx (I used data that come from Bruker preclinical animal scanner)
-
Biasfield correction of the B0 using ANTS N4biascorrection, and applied this correction to the whole 4D DWI dataset
-
Then, I ran the following tests for each individual shell
A-Response function calculations using the “tournier” algorithm using a mask containing whole cord at b-value 1500
dwi2response tournier < Input DWI> < Output response text file>
The above commands were repeated for the b=750.
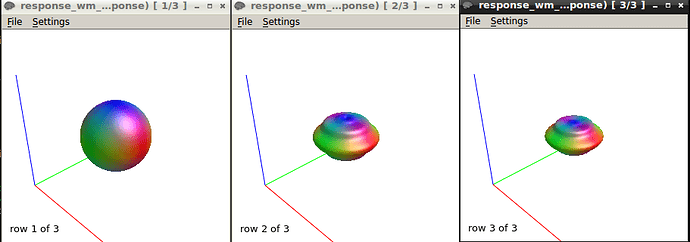
I got this response function at b1500.
The response function values at b1500 are
16616.92061510039 -3585.731063681506 662.1366725620043 -49.21846975544766 -372.193208900446 89.22703866558832
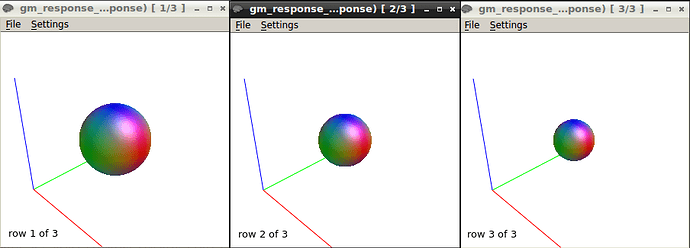
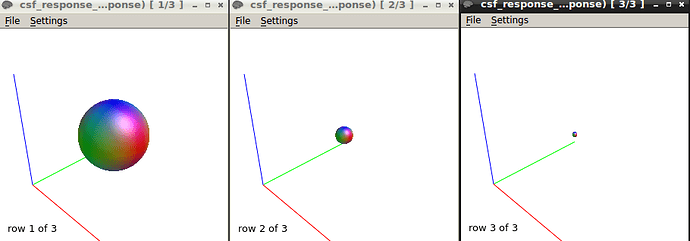
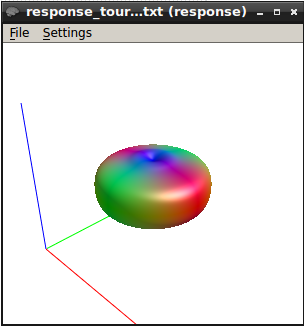
Also, I tested the TOURNIER algorithm on another sample with 2 shell (b0&3000). I got the following response function
The response function values at b3000 are 15261.69894075049 -3513.609487088777 1043.844357488741 -145.3679829867056 2.01888135870964 11.80721638417216
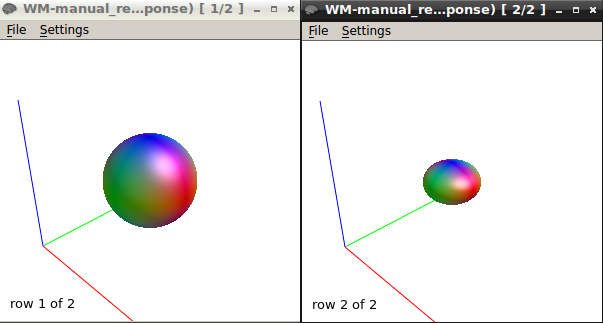
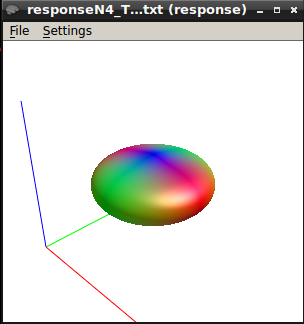
B-Response function calculations using the “tournier” algorithm using a mask containing selected voxels of SPINAL CORD WM AT b-value 750 & 1500
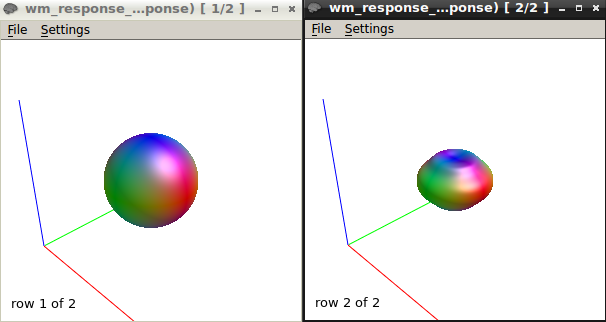
The response function values at b750 are
24174.5778079981 -1711.70084036846 442.5087763272201 13.27346114028632 12.25772995997397 -58.66570209134966
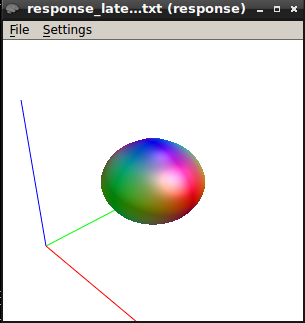
here is the response function using the same algorithm at b1500
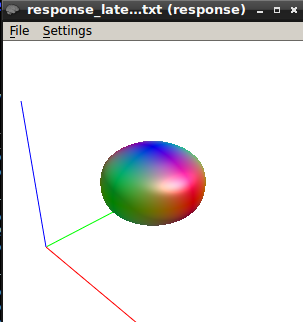
The response function values at b1500 are
9416.77124310872 -1993.300817125692 211.8305534782687 85.78772928903967 -89.50880832164782 54.91400798555078
*The other methods for response function result will be posted soon.
Many Thanks,
Hattan