Hi, everyone!
I’m trying to analyze the gliomas frequency in each imaging voxel. I first concatenated all the binary tumor mask(registered into MNI 512 space), and got the tumor frequency map(tumor_frequcency.nii). Then, I applied to the AAL3v1.nii and averaged the frequency voxel in each brain subdivision to obtain each subdivision frequency(tumor_frequcency_AAL3v1.nii). The AAL3v1.nii spacing is 222mm.
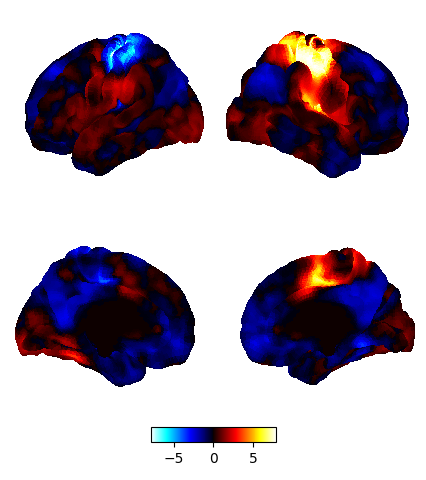
Now, I want to plot tumor_frequcency_AAL3v1.nii onto a surface. Firstly, I turn to the “nilearn” in python. The example is as below, but my result is stretched.
my result:
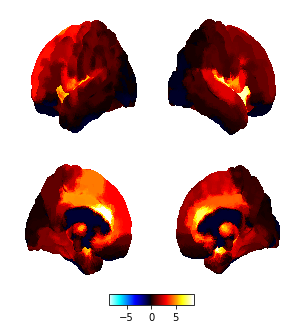
example:
here is my code:
tumor_map = nib.load(‘tumor_frequcency_AAL3v1.nii’).get_fdata()
fig = plotting.plot_img_on_surf(tumor_map,
views=[‘lateral’, ‘medial’],
hemispheres=[‘left’, ‘right’],
colorbar=True)
plotting.show()
I don’t know how to solve this problem. Given that the space of tumor_frequcency_AAL3v1.nii may be unmatch FreeSurfer’s fsaverage5(nilearn default surface)
, I think I may create the AAL3v1.nii surface by Freesurfer. However, it seems that it isn’t the usually way to relate anatomical template, AAL3, to Freesurfer, which more focus on the cortex pacellation.
I’ve read Problems with AAL parcellation and attempt to use command labelconvert to see whether it could help next.