Hi everyone. I am trying to restrict fixelcfestats to run in an roi-based manner, using a seed-to-target region that I generated from tckgen (using the population template). Simply swapping out the track ROI for the full tractogram in fixelcfestats does not work, probably because I’m still feeding it the whole-brain FD images and/or whole brain analysis masks. Maybe I need to max out height or extent? Or maybe there is a way to convert the tractogram ROI to a fixel analysis mask ROI ? I also tried using tcksample to create a track or vector file for each subject, but it’s not working :
tcksample tracks_20k_ROI.tck Subject1_fd.msf Subject1_fd_ROI.tck -force
tcksample tracks_20k_ROI.tck Subject1_fd.msf Subject1_fd_ROI.msf -force
The command seems to run, but when I try to open the resulting .tck or .msf files, I get exceptions:
mrinfo: [ERROR] unknown format for image "Subject1_fd_ROI.tck"
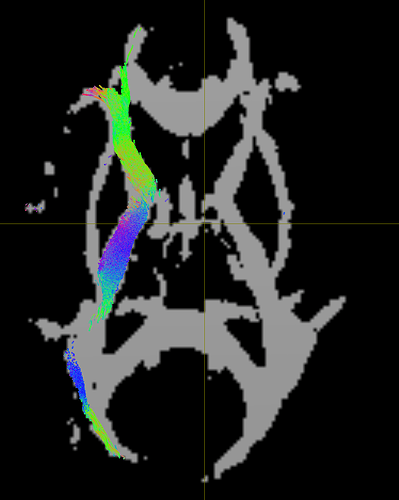
I have tried using other options in tcksample (-stat_tck, -precise) but I’m still getting the same error. For reference, here is the track ROI overlaid on Subject1_fd.msf:
In sum, I need advice on either: 1. restricting analysis to a tractogram-based ROI w/in fixelcfestats or 2. successfully creating ROI fixel maps for all subjects using tcksample.
Thanks in advance-happy to provide any other info.
Rachael