Hi Tournier,
Thanks for your detailed response, I really appreciate it and I can learn a lot from your response (& your contributions in this field).
That’s a pity I cannot reply immediately. Our server was down for several days, so I cannot get the data.
First of all, however, my problem is not a big one. There is no problem observed from mrview but ITK snap and more specifically, open segmentation button. This problem only takes place when I first open a T1 image and second open the label image.
Before my proble, I want say, I cannot use the operation ‘-copy_properties’ to fix this problem either, ROIs.nii.gz is the input.nii.gz, while the ROIs_test_strides.nii.gz is the output.nii.gz, and intensity-struct.nii.gz is the data before processing by Matlab - NIfTI package.
mrconvert 5tt/ROIs.nii 5tt/ROIs_test_strides.nii.gz -copy_properties mrbrain/intensity-struct.nii.gz
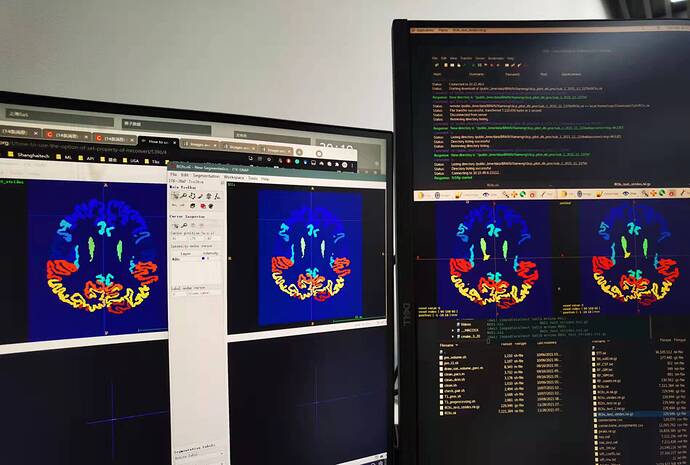
No matter in mrview or IT-SNAP, if I open the ROIs_test_strides.nii.gz and ROIs.nii, they look the same. as shown below. Sorry for the photo taken by my cellphone because the two tools are installed in two PCs.
mrconvert 5tt/ROIs_strides.nii.gz -set_property strides -1,2,3
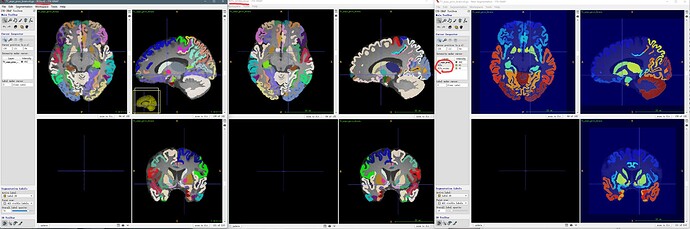
The figure below has three parts, the left two are the T1 image superimposed with label maps which name is marked by a red line, i.e., ROI.nii and ROIs_strides.nii.gz. Both are added to ITK-snap by the ‘open segmentation’ button. Meanwhile, the right one is opened by the ‘add another image button’. (Windows platform) And the right one demonstrate ROI.nii and ROIs_strides.nii.gz with no difference eighter.
So, here the problem only takes place when I first opened a T1 image and second opened the label image as a segmentation map.
As for the mrview, I think there is no problem, the fiber tracking results are well.
Left is ROI.nii and right is ROIs_strides.nii.gz.
This problem may not be a real issue, but I ask this question just for curiosity.
Thanks,
Feihong