Hello,
I have a question whether this method is fine for identifying the significant tracts from fixelcfestats results.
I have done the FBA analysis following the steps from the multi-tissue CSD tutorial (the only change is that I used SS3T-CSD with b=0 and b=2500). So, I have my significant results and I can display them either as fixels or on a tractogram Displaying results with streamlines — MRtrix 3.0 documentation. However, I would like to be able to identify the tracts with significant fixels. In order to do so I have done a TractSeg based tractography on the wmfod_template (used sh2peaks on the wmfod_template and run the tractography on the peaks file) and obtained the segmented tracts in .tck format. Next, and this is just an example of what I wanted to inspect, I used tckedit for combining left and right optic radiations and separetly left and right ILF.
tckedit tractseg_template/TOM_trackings/ILF_right.tck tractseg_template/TOM_trackings/ILF_left.tck ILF_all.tck
tckedit tractseg_template/TOM_trackings/OR_right.tck tractseg_template/TOM_trackings/OR_left.tck OR_all.tck
Then, I have used fixel2tsf to map the fixel derived 1-p values to selected tracts:
fixel2tsf stats_fdc/fwe_1mpvalue.mif ILF_all.tck stats_fdc/fwe_pvalue_ILF.tsf
fixel2tsf stats_fdc/fwe_1mpvalue.mif OR_all.tck stats_fdc/fwe_pvalue_OR.tsf
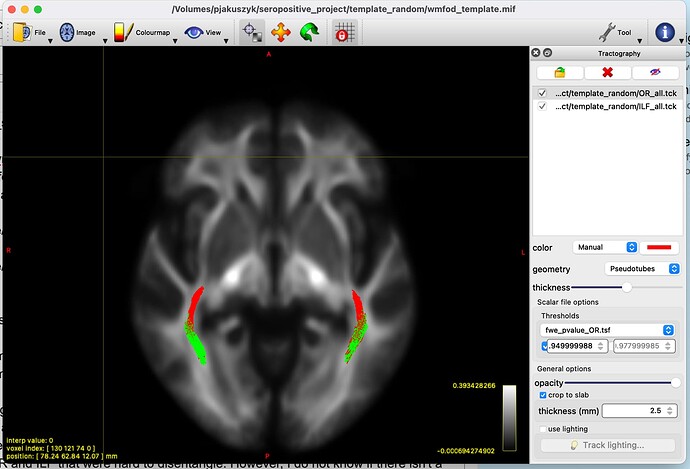
Finally, I use the same logic as in the manual to display the results. I load both the ILF_all.tck and the OR_all.tck to the mrview and threshold each by the appropriate file (fwe_pvalue_ILF.tsf, fwe_pvalue_OR.tsf respectively). The results are seem ok to me, I can tell that I have significant differences both in the OR and ILF that were hard to disentangle. However, I do not know if there isn’t a major flaw in doing something like that.
Thank you for your help,
Pawel
Picture of the final output (OR colored in red, ILF green):