My professor wants me to find slf fibres.
He gave me fod image.
My current codes are tckgen.exe FODfilename.mif TobemadeTckfilename.tck ROIfilename.mif -mask
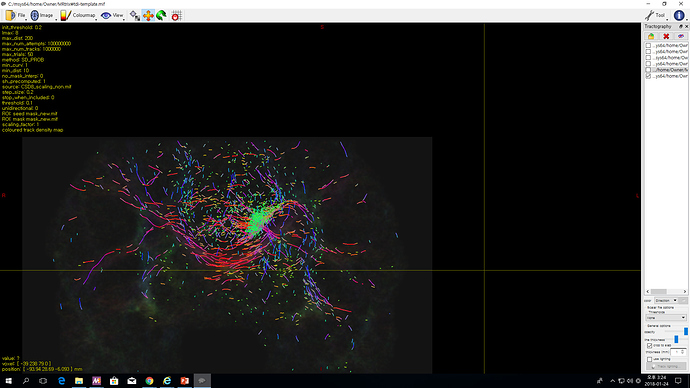
But the fibres tracked are no where near the sample that my professor showed me.
They are all randomly scattered over the FOD image, but they have to be stretching out neatly from the ROI point , right?
Can you give me some sample codes that i can try to improve on this?
Please help me
This would return an error and not run at all actually… I suspect you provided the ROI to the -include option? You could start by adding other include regions as an extra constraint (e.g. at both ends of structures you’re trying to track), or add exclude regions to get rid of some of the false positive spurious streamlines.
Also, when visualising the outcome, it may help to switch off “crob to slab” to get a better 3D impression of the whole result. It may very well be that the majority of your streamlines does represent (parts of) the core of the bundle you’re after, but at some point just randomly branches off. This may give a messy result, but it may not be “as bad” as it looks at first sight.
I’ve got the feeling you were given this as an assignment or exercise of sorts? If so, you’d best experiment a bit more around with the different options to tckgen that take ROIs (-include, -exclude, -mask, …). This will be the best way to get a good feeling about how you can achieve results with tractography in practice.
Thanks for the reply
Sorry, I think the code I’ve written on the question is different from what I’ve been using.
tckgen.exe FODfilename.mif TobemadeTckfilename.tck -seed_image ROIfilename.mif -mask Maskfilename.mif
Can you have a look at it and see what I can add on?
I think include exclude options that you have suggested are relevant when I’m not specifying ROIfile directly.
Thank you
Oh no, they’re definitely compatible with it. Essentially, now, you have a single seed region (and a mask); so naturally, your streamlines will start nicely there, and follow along your bundle of interest for a while, and even the majority will follow it for quite a distance… but eventually, streamlines will branch off here and there from the bundle, creating the mess of spurious fibres you see. Even though they’re all over the place, they’re still very sparse (and not dense at all). Adding exclusion regions will help you to eliminate some of these spurious streamlines, given that you put them in strategic locations of course. Extra inclusion regions will help you constrain the bundle to better go towards a particular target (along a particular orientation and path). To get the SLF, 2 well positioned inclusion regions (vaguely around the ends of the the SLF) will already get you much further. That, I’d do as a first step. Then look at that result, and refine it by adding exclusion regions where necessary, should any “stubborn” streamlines still take a different route from the one inclusion region to the other inclusion region.
But again, this takes some hands-on attempts, so you can gradually refine your command (and the regions you draw); and at the same time get the experience to better predict up-front how tractography will behave for particular tasks, such as tracing the SLF.
In case it helps: take a look at this paper, particularly Figure 5 and the associated text. It uses the previous version of MRtrix but the principles are the same…