Hello,
I previously rant ACT using the following commands however, when asked to clear up space on the linux system I use I removed the 10 million streamline tract files from the previous analysis. However, while I kept the connectome.csv output files, a few months later I needed to replicate/rerun tckgen and I accidentally overrode the assignment.csv and weight.csv file from the previous run. Meaning I can’t reproduce the tractogram from the previous analysis. This wouldn’t be a huge problem if the second time around the results were similar to the first analysis, however, my connectome.csv files appear to be quite different despite (I believe, a near-identical pipeline). Additionally, I know I have not altered the wmfod.mif and aparc+aseg.mgz file has not been modified since the first iteration. Additionally,I noticed I used the nodes_old.mif file instead of the nodes_fixSFM_old.mif file for tck2connectome the first time around so I kept that consistent this time around.
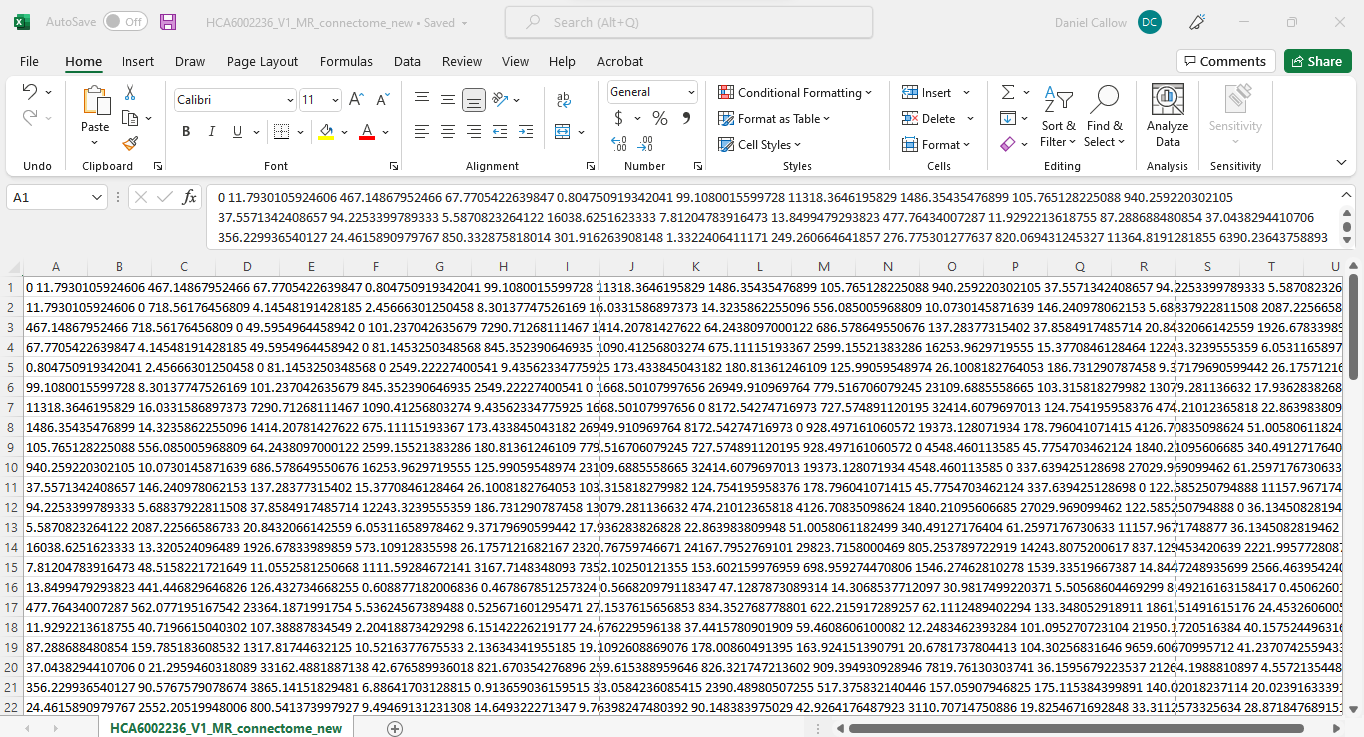
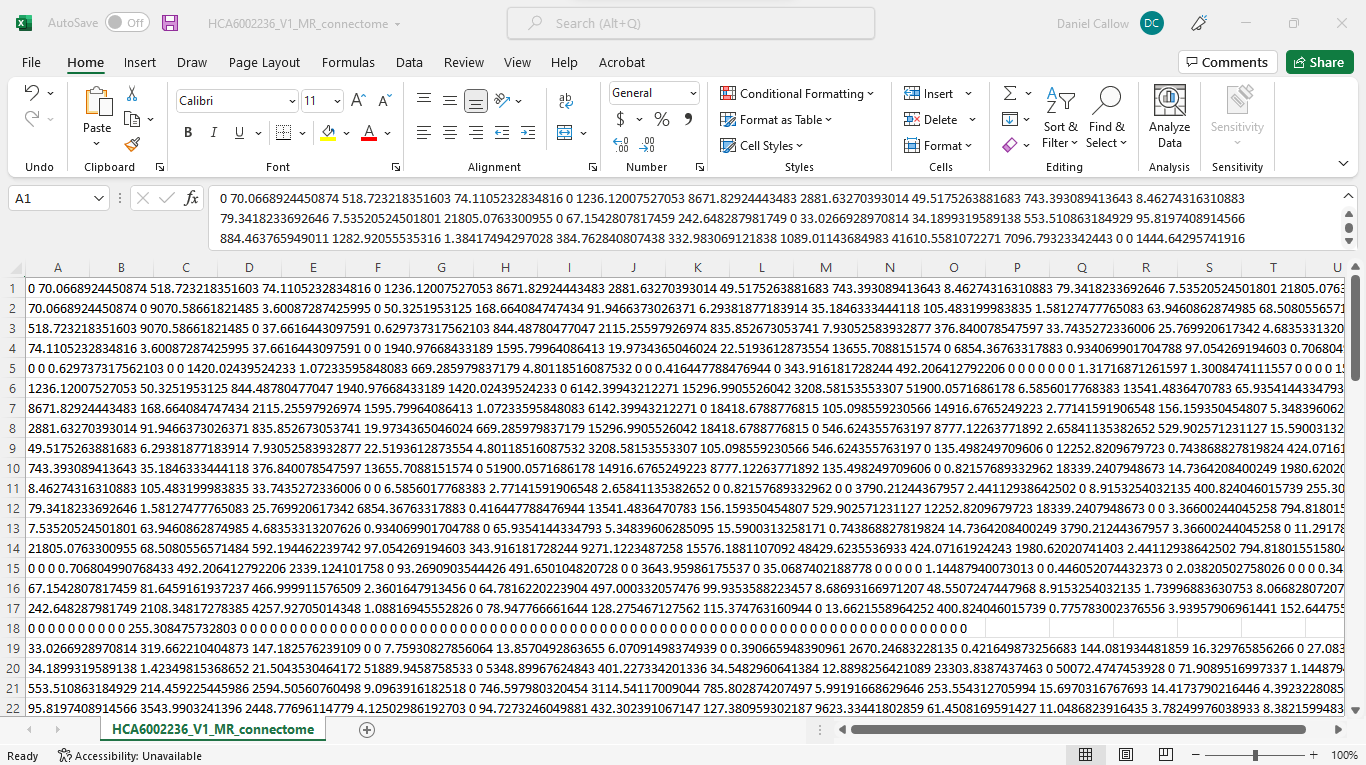
Attached are images of the new and old connectome (couldn’t fit the whole thing in an image unfortunately)
Below is the script I ran the second time and thought it was essentially identical to what I ran the first time. Just wondering if anyone might have an idea of what could be causing the differences or if this is just to be expected with -dynamic_seed. For reference, I think this is something else since the global and local efficiency measures I am getting are pretty drastically different between the two iterations.
First, I took the aparc+aseg file from freesurfer into diffusion space.
Then run 5ttgen with the freesurfer option
Convert and fix labels
and then run ACT with dynamic seeding, probabilistic tractography, and SIFT2
mri_vol2vol --mov lowb_brain_corrected.nii.gz --targ /data/bswift-1/dcallow/aging/freesurfer/${subj}/mri/aparc+aseg.mgz --inv --interp nearest --o cort_subcort.nii.gz --reg register.dat
5ttgen -sgm_amyg_hipp freesurfer cort_subcort.nii.gz 5TT.mif
labelconvert cort_subcort.nii.gz FreeSurferColorLUT.txt fs_default.txt nodes_old.mif
labelsgmfix nodes_old.mif cort_subcort.nii.gz fs_default.txt nodes_fixSFM_old.mif -premasked
tckgen -nthreads 4 -act 5TT.mif -backtrack -crop_at_gmwmi -seed_dynamic wmfod.mif -maxlength 250 -select 10M -step 1 -angle 45 -cutoff 0.06 wmfod.mif 10M.tck
tcksift2 -nthreads 4 10M.tck wmfod.mif -act 5TT.mif weights.csv
tck2connectome -force -nthreads 4 10M.tck nodes_old.mif connectome_old.csv -tck_weights_in weights.csv -out_assignments assignments_old.txt -symmetric -zero_diagonal
Thank you for any help or any suggestions on what I could look into/what could be leading to such different results!