Dear experts,
I’m currently working on building a multi-channel population template for rotator cuff muscles using FODs and mDixon anatomical scans from three subjects. This is part of a preliminary phase to test and optimize template parameters.
Issue Encountered: During the non-linear registration step of creating the population template, I’ve observed an anatomically implausible twist in the mDixon template, particularly around the scapula area. I’m trying to determine if this could be due to the use of the NCC metric or perhaps some suboptimal settings for NCC.
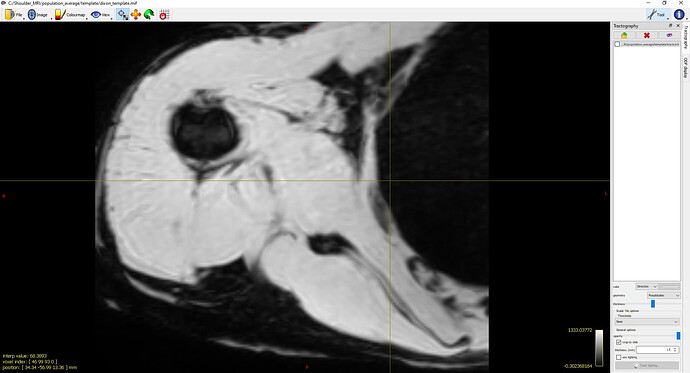
What I’ve Tried: To explore this further, I conducted two sets of tests with identical settings for rigid and affine registrations.The averaged mdixon images after rigid and affine registration appear satisfactory (refer to screenshot 1) :
However, the issue arises after the non-linear registration step. I’ve experimented with two different sets of settings for non-linear registration (the mask input contains segmentations for all four rotator cuff muscles):
- Settings A:
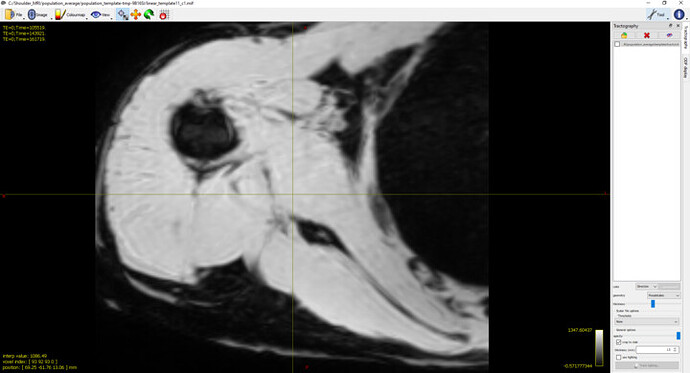
population_template ../template/fod_inout/ ../template/fod_template.mif ../template/dixon_input/ ../template/dixon_template.mif -mask_dir ../template/mask_input/ -template_mask ../template/mask_template.mif -voxel_size 0.75 -nl_metric_radius 3 -nl_disp_smooth 0.75 -nl_update_smooth 1 -nl_niter 50,50,50,50,50,50,50,50,50,50,50,50,50,50,50,50(See Screenshot 2)
- Settings B:
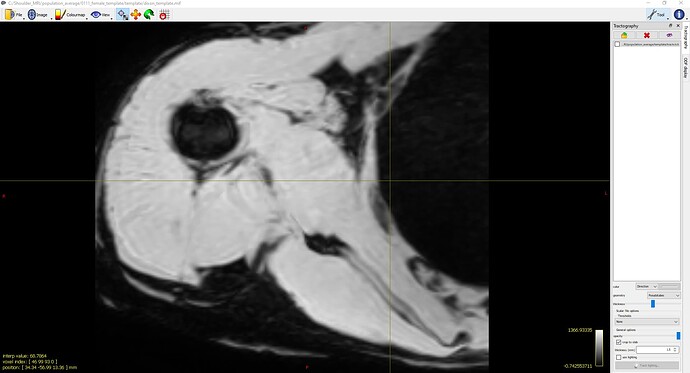
population_template ../template/fod_inout/ ../template/fod_template.mif ../template/dixon_input/ ../template/dixon_template.mif -mask_dir ../template/mask_input/ -template_mask ../template/mask_template.mif -voxel_size 0.75 -nl_metric_radius 5 -nl_disp_smooth 0.75 (default 1.0 x voxel_size) -nl_update_smooth 1.5 (default 2.0 x voxel_size) -nl_niter 5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5(See Screenshot 3)
While the second setting slightly reduced the twisting, it was still noticeable. Interestingly, when I switched to SSD for non-linear registration, the twisting issue was resolved.
Seeking Insights: Could this issue be related to a specific non-linear registration algorithm used in MRtrix? Any suggestions for adjusting the settings or insights into the potential cause of this issue would be greatly appreciated.
Quantitative Evaluation Methods: Lastly, I’m also brainstorming methods to quantitatively evaluate the registration and template-building process. My current ideas include:
- Comparing the Dice coefficient for muscle labels and angular coefficient between FODs.
- Warping the FOD template back to the subject’s space and calculating the angular coefficient between the ground-truth FOD (derived directly from the subject’s DWI) and the warped FOD template.
I would be grateful for any suggestions or additional methods that the community might recommend for this evaluation.
Thank you in advance for your time and assistance!
Best regards,
Yilan