Hi guys,
I am trying to build some age-specific brain templates using DW data from a public database and I need your help with some issues that emerged during the processing:
-
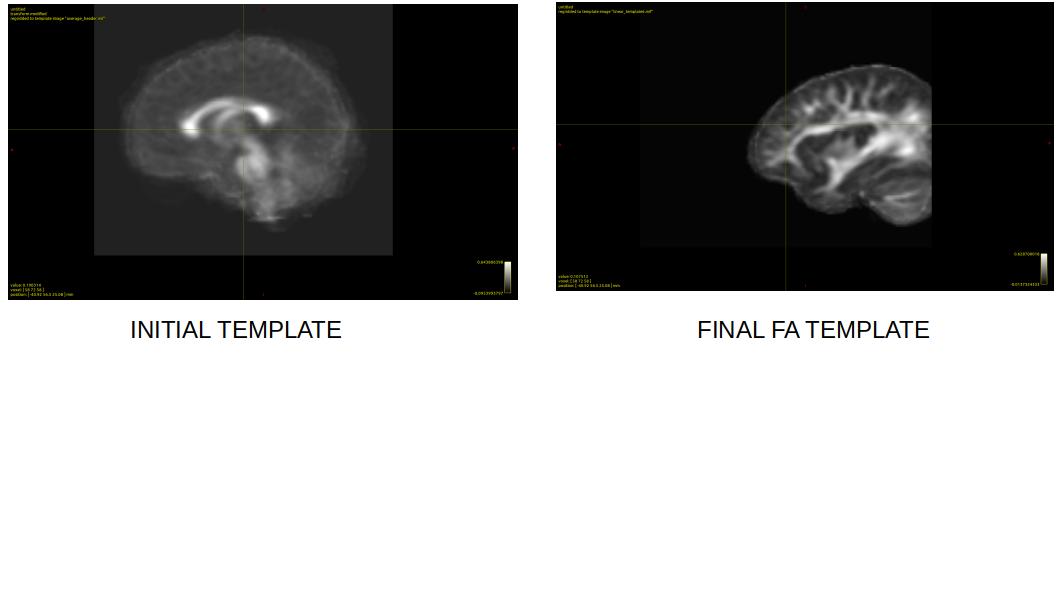
When I run dwiintensitynorm to normalize signal intensities across subjects and to create a mean FA template, the output FA template is cropped (see figure)
-
When I run population_template to create a FOD template, the system crashes. Here is the error:
population_template: [100%] Erasing temporary mask images
population_template: [ 0%] Performing initial rigid registration to template... population_template: [ERROR] Command failed: mrregister /isilon/data/filippo.arrigoni/provatemplatemrtrix0305/mrtrix/FOD/1442_V3_FOD.mif average_header.mif -mask1 /isilon/data/filippo.arrigoni/provatemplatemrtrix0305/mrtrix/MASKS/1442_V3_dwi_bc_ups_mask.mif -rigid_scale 1 -rigid_niter 0 -type rigid -noreorientation -rigid_init_translation mass -datatype float32 -rigid linear_transforms_initial/1442_V3.txt (population_template:492)
population_template: Output of failed command:
mrregister: [ERROR] Error allocating memory for scratch buffer
population_template: Script failed while executing the command: mrregister /isilon/data/filippo.arrigoni/provatemplatemrtrix0305/mrtrix/FOD/1442_V3_FOD.mif average_header.mif -mask1 /isilon/data/filippo.arrigoni/provatemplatemrtrix0305/mrtrix/MASKS/1442_V3_dwi_bc_ups_mask.mif -rigid_scale 1 -rigid_niter 0 -type rigid -noreorientation -rigid_init_translation mass -datatype float32 -rigid linear_transforms_initial/1442_V3.txt
Do you think that this is related to the storage resources of my workstation or to something else?
- Is there a way to visualize/open RGB maps generated by DTITK with MRVIEW? I bult a DTI template with DTITK, obtaining both FA and RGB mean images. The FA map can be opened with MRVIEW, whilte the RGB file can’t be properly visualised
Thank you for your time and help.
Best,
Filippo