Hi all,
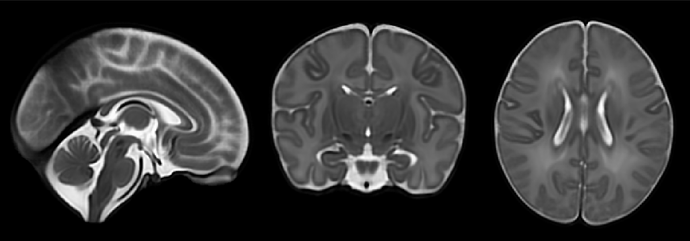
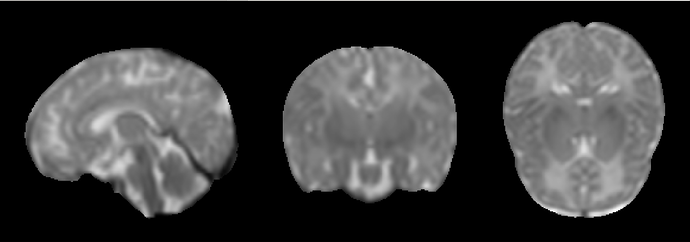
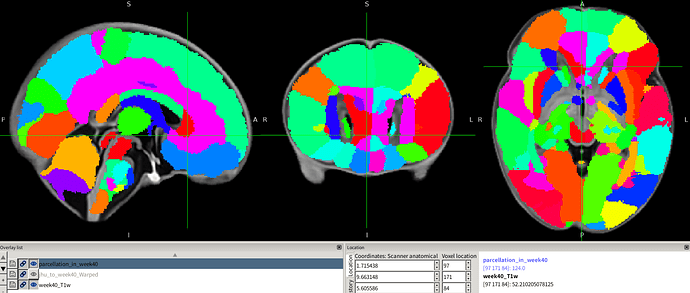
I want to register the JHU_atlas_SS_parcellation.nii.gz (Figure 1) to the space of my T2_template.nii.gz (Figure 2). My approach was to use FSL or ANTs to register the JHU_atlas_SS_t2ss.nii.gz (Figure 3) to the T2_template.nii.gz space, then apply the resulting deformation field to the parcellation atlas. However, the outcome (shown in Figure 4) is not satisfactory. My initial speculation is that this may be due to the low resolution of the JHU_atlas_SS_t2ss.nii.gz compared to my T2_template.nii.gz.
I would like to know if there are alternative directions/methods to achieve accurate registration of the JHU neonate WM atlas to my T2 image space.
Best Regards,
Hanjing
Hi,
It looks like the dHCP template right? What command did you use?
The authors of this atlas have a new tool for parcellation of the neonatal brain using the same protocol as the JHU atlas (they tested it with diffusion images), as this is a high resolution template, you could try it an see what happens.
Best regards,
Manuel
Yes, the T2 template is indeed the dHCP 40-week template (week40_T2w.nii.gz ). Here’s the exact ANTs command I used for registration:
# Paths
moving_image="JHUneonate_t2ss.nii.gz"
fixed_image="week40_T2w.nii.gz"
parcellation="JHUneonate_130parcellation.nii.gz"
# ANTs registration pipeline
antsRegistration --dimensionality 3 \
--float 0 \
--output [jhu_to_week40_,jhu_to_week40_Warped.nii.gz] \
--interpolation Linear \
--winsorize-image-intensities [0.005,0.995] \
--use-histogram-matching 0 \
--initial-moving-transform [$fixed_image,$moving_image,1] \
--transform Rigid[0.1] \
--metric MI[$fixed_image,$moving_image,1,64,Regular,0.25] \
--convergence [1000x500x250x100,1e-6,10] \
--shrink-factors 8x4x2x1 \
--smoothing-sigmas 3x2x1x0vox \
--transform Affine[0.1] \
--metric MI[$fixed_image,$moving_image,1,64,Regular,0.25] \
--convergence [1000x500x250x100,1e-6,10] \
--shrink-factors 8x4x2x1 \
--smoothing-sigmas 3x2x1x0vox \
--transform SyN[0.05,3,0] \
--metric CC[$fixed_image,$moving_image,1,4] \
--convergence [200x150x100x50,1e-6,10] \
--shrink-factors 10x6x4x2 \
--smoothing-sigmas 4x3x2x1vox
# Apply transforms to parcellation
antsApplyTransforms -d 3 \
-i $parcellation \
-r $fixed_image \
-o parcellation_in_week40.nii.gz \
-n NearestNeighbor \
-t jhu_to_week40_1Warp.nii.gz \
-t jhu_to_week40_0GenericAffine.mat