Hi,
I am battling tracking on HCP dataset with particular methods from dipy (e.g. CSA Peaks, Maximum Direction Getter). I am assuming that something really fishy is happening to the masks because streamlines at the end seem to start… outside the brain. Perhaps due to a small experience, I am doing something wrong on the way. I would really appreciate it if someone can review my approach. ![]()
HCP dataset
DTI: 1.5 mm
T1: 1mm
Coregistration and masks generation
For my specific approach I need to coregister T1 to DTI, which I am doing as follows:
flirt -in ./anat/T1/T1.nii.gz -ref ./diff/raw/mri/diff.nii.gz -out ./tracking_files/t1_coreg_to_diff.nii.gz -omat ./tracking_files/t1_coreg_to_diff.mif -dof 6
5ttgen fsl ./tracking_files/t1_coreg_to_diff.nii.gz ./tracking_files/5TT_coreg_to_diff.mif -nocrop -nthreads 6
mrconvert -coord 3 2 -axes 0,1,2 ./tracking_files/5TT_coreg_to_diff.mif ./tracking_files/wm.nii.gz
5tt2gmwmi -nthreads 10 ./tracking_files/5TT_coreg_to_diff.mif ./tracking_files/wmgm.nii.gz
Mask for GMWMI of a functional region
Now that I have T1 coregistered I want to get a mask of GMWMI of a particular functional region. For that purpose, I go to FSLEeyes and use standard data in MNI152 space, get the functional region masks that are saved as:
BA44_L_thr20.nii.gz
BA44_R_thr20.nii.gz
These are binarized masks, threshold value of 20.
Now I want to move them from MNI152 space to a subject space. For that I am following this tutorial from mrtrix using prepared bash script:
export templateImage="/usr/local/fsl/data/standard/MNI152_T1_1mm_brain.nii.gz"
export templateImageMask="/usr/local/fsl/data/standard/MNI152_T1_1mm_brain_mask.nii.gz"
export subjectT1=$1
export masksDIR=$2
echo ${masksDIR}BA44_L_thr20.nii.gz
mkdir mask_temp
echo " "
echo "Extracting brain from T1 scan"
echo " "
bet2 ${subjectT1} ./mask_temp/T1_brain -m
echo " "
echo "Calculating transforms and masks"
echo " "
mrcalc ${subjectT1} ./mask_temp/T1_brain_mask.nii.gz -mult ./mask_temp/T1_masked.nii.gz
mrcalc ${templateImage} ${templateImageMask} -mult ./mask_temp/template_image_masked.nii.gz
flirt -ref ./mask_temp/template_image_masked.nii.gz -in ./mask_temp/T1_masked.nii.gz -omat ./mask_temp/T1_to_template.mat -dof 12
echo " "
echo "Calculating dilated transforms, masks and warp"
echo " "
maskfilter ./mask_temp/T1_brain_mask.nii.gz dilate - -npass 3 | mrconvert - ./mask_temp/T1_brain_mask_dilated.nii.gz -strides -1,+2,+3
maskfilter ${templateImageMask} dilate - -npass 3 | mrconvert - ./mask_temp/template_mask_dilated.nii.gz -strides -1,+2,+3
fnirt --config=T1_2_MNI152_2mm.cnf --ref=${templateImage} --in=${subjectT1} --aff=./mask_temp/T1_to_template.mat --refmask=./mask_temp/template_mask_dilated.nii.gz --inmask=./mask_temp/T1_brain_mask.nii.gz --cout=./mask_temp/T1_to_template_warpcoef.nii
echo " "
echo "Getting inverse of a warp"
echo " "
invwarp --ref=${subjectT1} --warp=./mask_temp/T1_to_template_warpcoef.nii --out=./mask_temp/template_to_T1_warpcoef.nii
echo " "
echo "Applying warp to a mask file"
echo " "
applywarp --ref=${subjectT1} --in=${masksDIR}BA44_L_thr20.nii.gz --warp=./mask_temp/template_to_T1_warpcoef.nii --out=./mask_temp/BA44_L_thr20_subj_space.nii.gz --interp=nn
applywarp --ref=${subjectT1} --in=${masksDIR}BA44_R_thr20.nii.gz --warp=./mask_temp/template_to_T1_warpcoef.nii --out=./mask_temp/BA44_R_thr20_subj_space.nii.gz --interp=nn
echo " "
echo "Mixing masks"
echo " "
mkidr masks
mrcalc ./mask_temp/BA44_L_thr20_subj_space.nii.gz ./mask_temp/BA44_R_thr20_subj_space.nii.gz -max ./mask_temp/BA44_RL_thr20_subj_space.nii.gz
mrcalc ./mask_temp/BA44_RL_thr20_subj_space.nii.gz wmgm.nii.gz -and ./masks/BA44_wmgmi_thr20.nii.gz
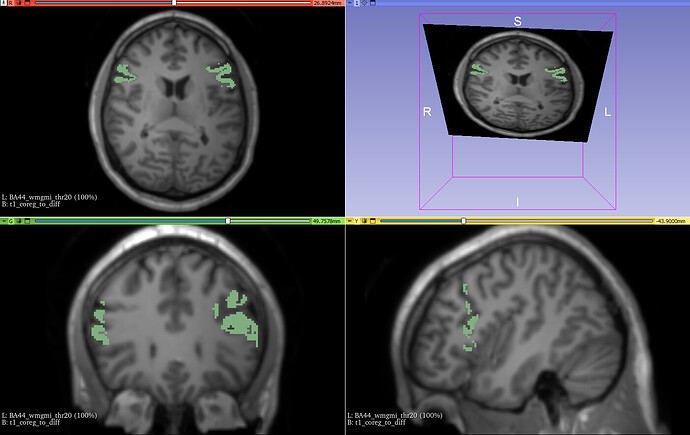
Now comes the weird part. When I visualize mask on a T1 volume coregistered with T1 everything looks ok in Slicer:
However, when I throw in diffusion data (diff.nii.gz) the mask is really off:
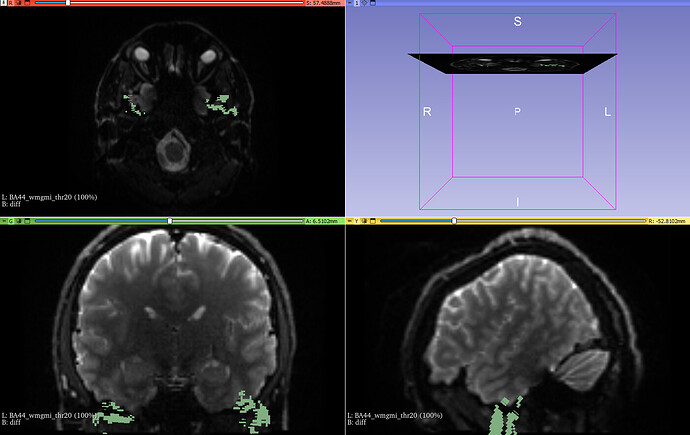
Now the weird thing is when I visualize diffusion data with a mask in FSLEyes it seems to be ok:
although FSLEyes shows the message in location pane:
Displaying images with different orientations/fields of view!
BA44_wmgmi_thr20
[39 93 47]: 0.0
diff
[100 46 47 0]: 666.577392578125
It seems then the coregistration is off?
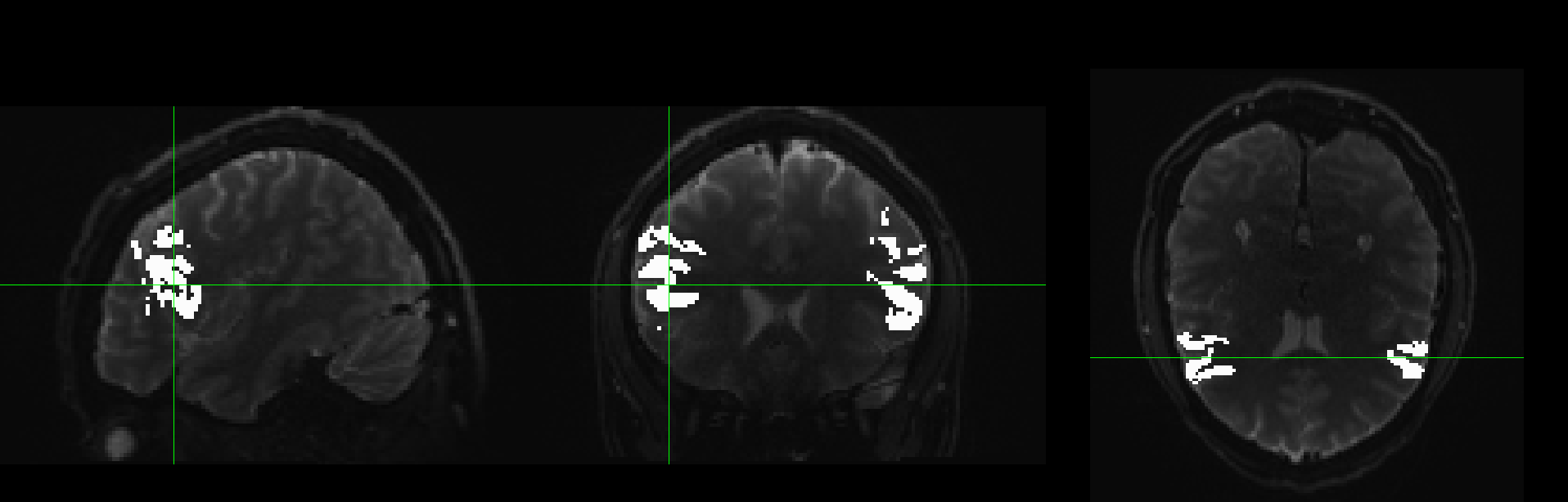
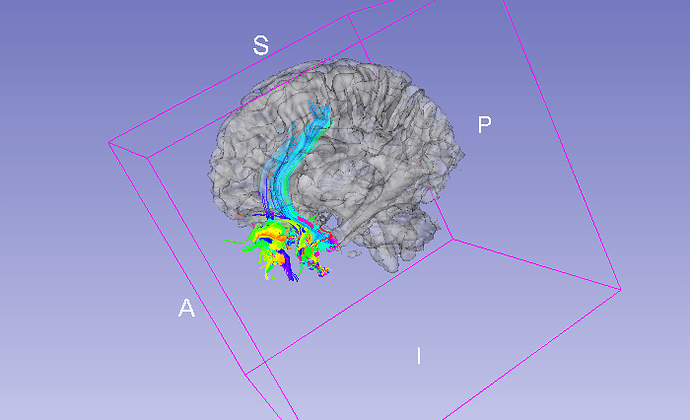
When I try to track from this setup, using mask shown here as a source of seeds, it seems that it is really off:
It seems that tracking really started in a place indicated by mask overlayed with diffusion in slicer! How come?
I thought maybe I just need to transform the streamlines, so I did, using DTI affine:
from dipy.io.image import load_nifti
from dipy.tracking.streamline import transform_streamlines
data, aff, img = load_nifti(arguments.dti_reference, return_img=True)
streamlines = transform_streamlines(tractogram.streamlines, aff)
but then it was even more off and transform of a inverse affine did not help as well:
streamlines = transform_streamlines(tractogram.streamlines, np.linalg.inv(aff))
Am I doing here something fundamentally wrong?
Best regards,
Mateusz