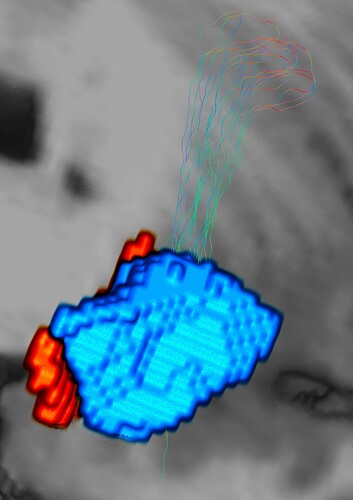
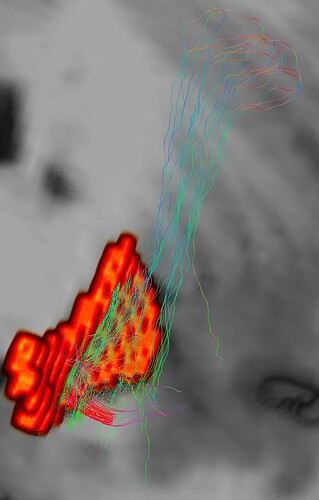
Hi guys, thank you for taking time to help me out. I have generated a whole brain tractography using tckgen and edited the tracts between anterior hippocampus and entorhinal cortex. These two masks are adjacent to each other and I was expecting to see tracts that connect these two masks. I also created an exclusion mask to make sure the output only include my interested tracts.
But instead of the expected straight-forward tracts, the output came up weirdly with extra tracts looped around the hippocampus then arrived at the entorhinal cortex. The ends_only command was able to show this loop tracts more clearly. One of the experts in hippocampus told me that this is error caused by poor image quality and there is no way to fix it. I know the possibility is low but i still want to give it a shot.
Hope these images make sense. The blue mask is anterior hippcampus and the red is the entorhinal cortex. And there are several loops at the posterior hippocampus part.
Would it be possible to remove those looping streamlines and only leave the right ones which are the curved red streamlines. Thank you in advance.