Dear experts,
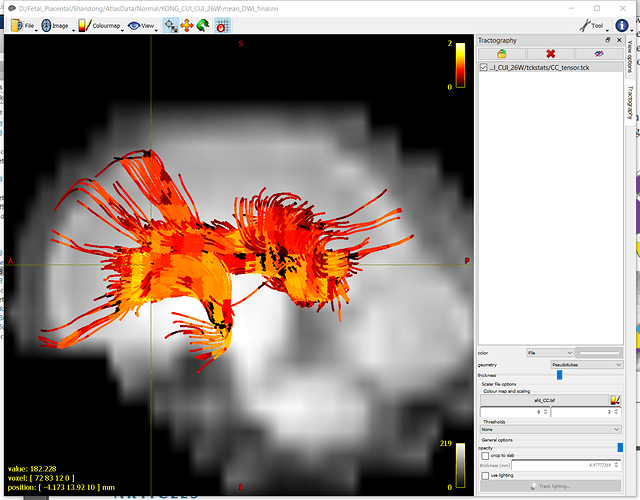
I am trying to map scalar values (fa.nii, mk.nii, etc.) onto the tracts for visualization. I realize .tsf files can be loaded and mapped onto tracts like the screenshot below, but is there a way to map conventional 3D images onto the tracts?
Thanks a lot!
Dan Wu