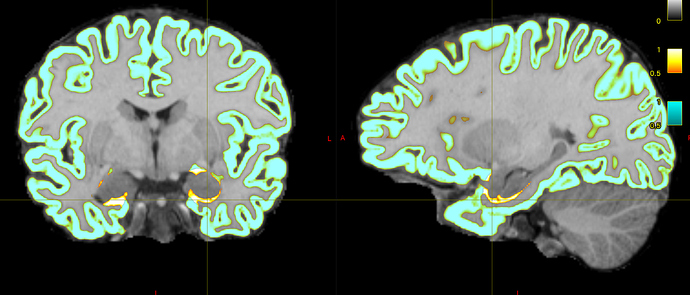
I initially used -hippocampi first , and noticed a few voxels adjacent to the amygdala and hippocampus that are still labelled as cortical grey matter
I believe that I have observed similar in some circumstances myself. The issue is that the FreeSurfer white and pial surfaces do in fact separate from one another spatially at that location, and therefore in the process of mapping those two surfaces to partial volume images and using their difference as the cortical GM fraction there does end up being some erroneously-segmented voxels. What I’d need to do to fix this is to additionally read from the binary vertex masks generated by FreeSurfer to indicate which vertices are cortex and which are “false” vertices generated in order to provide closed surfaces, and modify the surface-to-image conversion process to take this into account, which is unfortunately quite clumsy and requires a decent amount of change. It would make my own life a lot easier if the two FreeSurfer surfaces simply didn’t diverge from one another spatially where there’s no cortex… 
When comparing this with the -hippocampi aseg flag, its easy to see that FIRST underestimates these regions relative to FreeSurfer.
I wouldn’t say “under-estimates”; personally I trust the FIRST segmentations moreso than aseg. “Estimates a smaller volume” maybe?
Also note that if you use the FreeSurfer hippocampal subfields module, 5ttgen hsvs will utilise its output. That’s probably the preferable of the three.
So I’m trying to understand how this issue will ultimately affect tckgen and tck2connectome. … If so, is it better to use aseg?
As long as the thickness of that erroneous “cortical” GM band is less than the maximum distance in the tck2connectome radial search maximum distance, streamlines that should actually be intersecting the hippocampus will still be assigned as such in either case, even if in one instance streamlines may project into the hippocampus whereas in the other they are terminated immediately. If there are any pathways that run parallel to the surface between the hippocampus and the WM, then there’s a chance that those streamlines may terminate prematurely in the former case whereas in the latter they will continue further through the WM.
And then I started wondering whether my choice of amyg/hippo segmentation in 5ttgen needs to align with the parcellations provided to tck2connectome. In other words, if I use -hippocampi first , do I need to use labelsgmfix -sgm_amyg_hipp ?
It’s preferable for the tissue segmentation for ACT and the parcellation for tck2connectome to not have gross discrepancies between them. The default assignment mechanism in tck2connectome does tolerate some mismatch at the cost of some false positives in other scenarios, but if you can use the same source of information for segmentation of a particular structure that’s almost certainly going to be preferable than using different ones. This is essentially the result shown in the appendix of this manuscript, which is where labelsgmfix came from.
Cheers
Rob