Hello,
I’ve come across two peculiar issues with the fixel-based pipeline.
The first is with the resultant voxel size from population_template. I see that it looks through all the image headers (via mraverageheader) to create the final template image. With that being said, the resultant voxel size does not match those of what is expected across the input images.
For example, I have inputted all FOD images with an upsampled voxel resolution of 1.5 mm (stride +1,2,3,4) cubic squared. However, the resultant voxel dimensions are 1.63718 x 1.73726 x 1.72173 mm, with the strides now [ 2 3 4 1 ].
I’ve doubled checked the image headers (of the upsampled FODS), and all image headers have voxel resolution of 1.5.
One more thing.
Across my single patient cohort - I’ve extracted the “fixels” with a peak of 0.33 - resulting in 71900 voxels for subsequent analysis.
I’ve looked at the fdc amplitudes across the subjects, and a lot of individuals are not consistently exhibiting non-zero values for the initial voxel-populations (i.e. the first 80 or so voxels).
Here i’ve represented the log of the fdc values, for better visualisation:
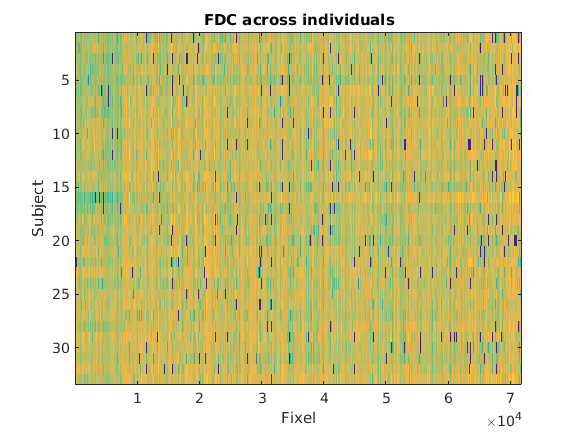
This is merged with the above issue as I believe the spatial resolution/reorientation could make the fd/fc values to be calculated outside of the brain tissue?
I was wondering whether there was a way to determine the spatial location of the above-threshold fixels? The resultant fdc.mif images are in the dimensions [71900 1 1], and hence are not easily able to visualize.
Of course, this is a heterogeneous clinical population (Parkinson’s), and perhaps these zero fdc values are not surprising.
Again @rsmith, you can check my github repos for further information.
Best,
Alistair