Dear MRtriX Team,
I have a question concerning the mrconvert command. I got different outputs when I convert my DWI data from dicom to mif directly compared to dicom to nifti and then via fsl_grad to mif?
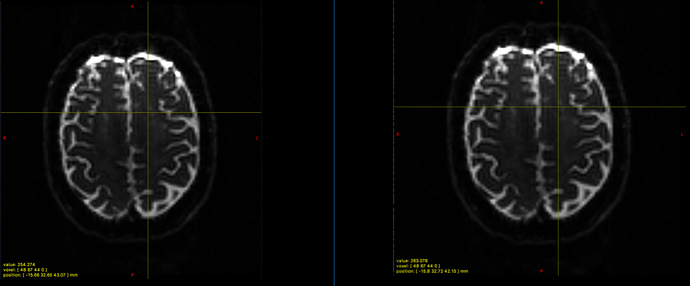
The scanner coordinates are slightly different and also the intensity value (see picture 1).
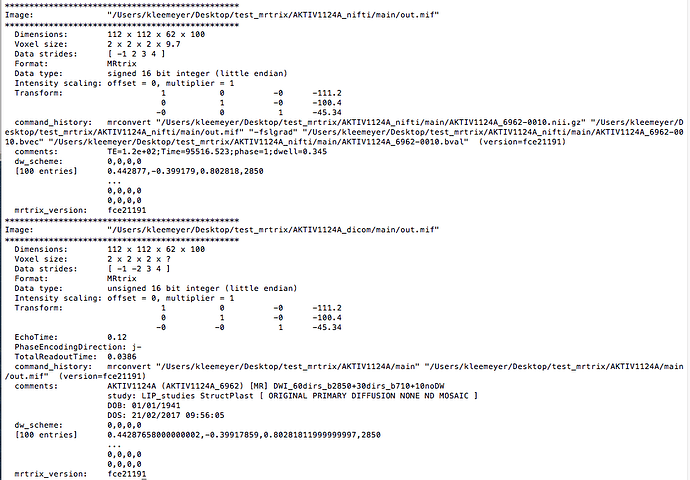
Furthermore, the header information are different (see picture 2 strides).
According to my processing steps I need also the nifti files, however I am not sure which are correct, since they the outputs are different.
Did I something wrong?
Thank your for your support!
Best
Max
Picture 1
Picture 2
I don’t think there’s anything to worry about here:
-
For the first set of images, remember that the ‘position’ entry will report the exact location of the crosshairs in 3D space, while the ‘voxel’ entry will give you the integer voxel indices of the corresponding voxel. So you can be in the same voxel, but at slightly different locations (as long as they’re both within the voxel, of course). Also, when image interpolation is on (the default, and on in your case), the ‘value’ entry shows the value obtained by trilinear interpolation at the position of the crosshairs - not at the nearest voxels. However, when interpolation is turned off (View->interpolation, or shortcut key I), then the ‘value’ field will be that of the nearest voxel. Hopefully you’ll find that values match again in that case.
-
Your mrinfo output shows agreement on everything but the ~6th decimal place of the DW gradient direction. That’s not unexpected, given that this information needs to be manipulated differently for the two formats, and the values involved in the calculation are typically stored as 32 bit floats (for the NIfTI format), and 6 decimal places is about the level of accuracy you’d expect for 32 bit floats…
Thank you for your quick reply,
May I ask you one more question about the mrinfo output.
Why are the data strides different? -1 2 3 4 vs. -1-2 3 4 and what does the voxel size 2 x 2 x 2 x 9,7 vs. 2 x 2 x 2 x ? mean?
Thank you
Best
Max
I’m not sure, that should match. At least it does on my system:
$ mrconvert dicom/ dwi.mif
$ mrconvert dicom/ dwi.nii
$ mrconvert dwi.nii dwi2.mif
$ mrinfo dwi.nii dwi2.mif -stride
-1 -2 3 4
-1 -2 3 4
I expect you’ve set the strides manually in your conversion to NIfTI? Was it even performed within MRtrix3, or using some other 3rd party converter? Many of them actually set the strides explicitly to something that makes interaction with e.g. FSL easier.
It means the spacing between voxels is 2 mm in the spatial axes, and 9.7 along the volume axis, which is presumably the repetition time in units of seconds. This would have been read directly from the DICOM headers by whatever software was used for the conversion (clearly not MRtrix3, by the sounds of it – guess that answers my previous question). In the direct DICOM conversion via MRtrix3, the spacing along that axis is left undefined (hence the ? for final value), since there is no natural ‘spacing’ between volumes in a DWI series. Other conversion tools might store the TR there as this makes sense in applications like fMRI.
Thank you for your answer,
I used the tool. dcm2nii . For the next calculations I will just do it via
$ mrconvert dicom/ dwi.nii as you suggest.
Thank very much!
Best
Max
No need to use mrconvert if you’re comfortable with dcm2nii, there’s nothing wrong with it. It might convert slightly differently, but the results are equally valid, and MRtrix3 will have no trouble using them. I was just answering your questions as to why there was a difference, not suggesting that one is better than the other…