Hi everyone,
I am running Mac OS BigSur 11.2.3 on Intel i9 8-core 2.4 GHz, 64GB memory. I have installed both MRTrix3 and MRTrix 3Tissue, but only MRtrix3 is exported to my $PATH.
I am trying to register a subject’s T1 scan to their diffusion scan with the following commands:
flirt -in $DIFFUSION -ref $T1 -dof 6 -cost normmi -omat dwi2T1.mat -out dwi2T1.nii
convert_xfm -inverse -omat T12dwi.mat dwi2T1.mat
transformconvert dwi2T1.mat $DIFFUSION $T1 flirt_import dwi2T1_mrtrix.txt -force
mrtransform $t1 -linear dwi2T1_mrtrix.txt -inverse T12dwi.nii -force
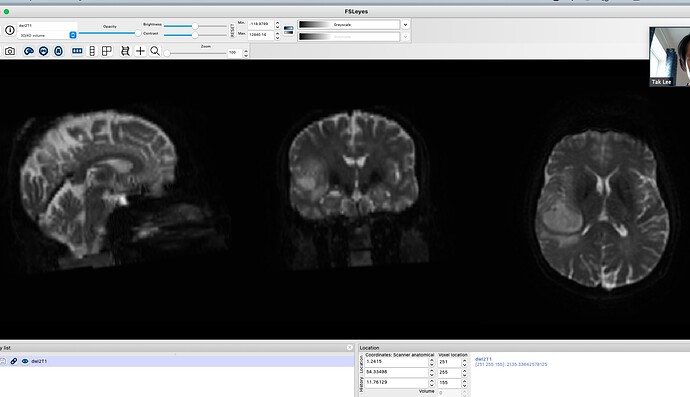
The problem is that my final output T12dwi.nii image contains missing slices intermittently when I run the above commands. No error is thrown and it can work but if I run the same commands again- the displayed image shows a slice or 2 missing- see attached image.
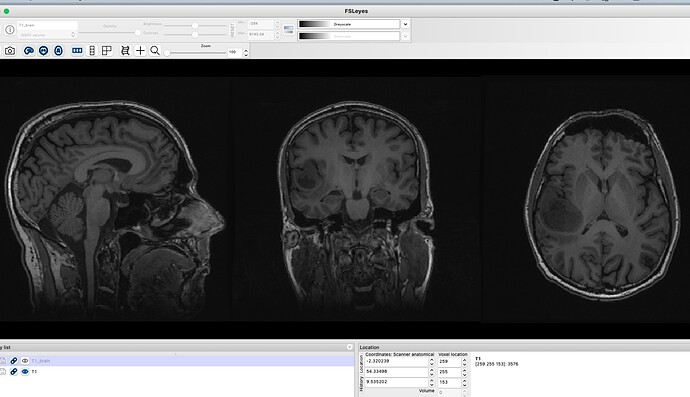
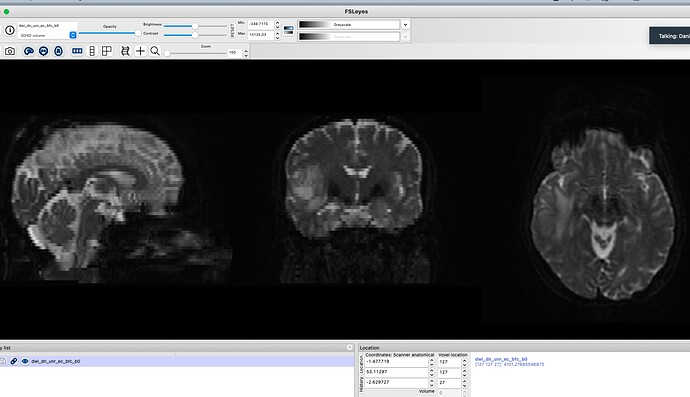
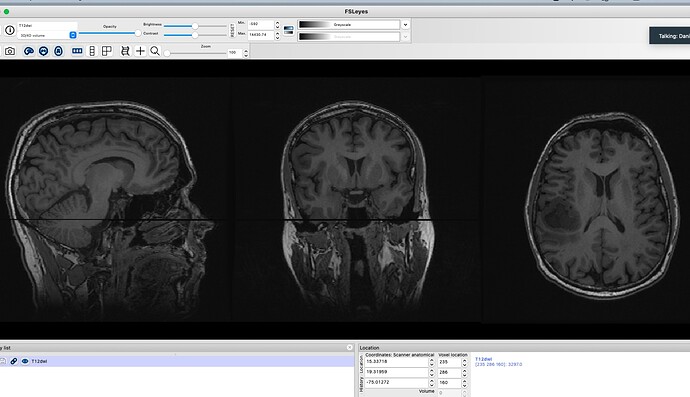
I am unsure why this is and it does not seem to be rendering issue as this is seen in mrview and fslview. I know that my input T1 and DIFFUSION scan are fine- see attached. I also know that dwi2T1.nii from the flirt output is also fine- see attached.
The diffusion scan has been preprocessed:
mrdenoise → mrdegibb → fsl TOPUUP (TOPUP was run after synb0 as I only have 1 b0: GitHub - MASILab/Synb0-DISCO) → fsl EDDY → dwibiascorrect → mrconvert -coord 3 0 -axes 0,1,2 to extract b0 image (I only have 1 b0 image).
Has anyone encountered this before?
Thanks!
Yizhou
Hi Yizhou,
Is $t1 the same as $T1? (BTW, you are not using the output of convert_xfm.)
mrtransform -linear without for instance the -template option does not modify the voxel data, the transformation is applied to the image header. So unless the dark stripe is visible in the input (mrview $t1 -interpolation 0 or $T1), I have no idea what might be causing this issue. To get to the bottom of this:
- Is this stripe pattern reproducible also with
mrtransform $t1 -linear dwi2T1_mrtrix.txt -inverse T12dwi_0thr.nii -nthread 0?
- If your working directory is not local, does the problem occur when you write
mrtransform’s output to /tmp/T12dwi.nii instead?
- Can you reproduce the output of
mrtransform using the same data and transformation on another computer?
- What is the value in the affected slice, is it zero or NaN? You can use mrview’s crosshair with image interpolation switched off.
- What version of MRtrix3 are you using (
mrtransform -version)?
- What is the output of
mrtransform $t1 -linear dwi2T1_mrtrix.txt -inverse T12dwi_debug.nii -debug
- What is the content of
dwi2T1_mrtrix.txt?
Cheers,
Max
Hi Max,
Thanks for your answer.
I have tried to reproduce the problem but it seems to have disappeared… I was unable to recreate the problem on a different computer. I then modified my pipeline to run 5ttgen on my T1 prior to running flirt and mrtransform to bring the 5tt output in DWI space.
Thus far, it seems to have worked. If it happens again, I will try your detailed suggestions and update the forum with the outcome.
Thanks again for the suggestions.