Hello all,
I was just curious to see if anyone has performed tractography on images obtained via MAP (mean apparent propogator) MRI. Is it even possible? If so, what’s the general workflow like. Any insight would be great! Thank yall in advance.
Cheers,
Ricardo
If you acquired your data as in the original MAP MRI paper then it is just a multi-shell sequence (with many b-values and a very high maximum b-value). So you should be able to follow our “standard” multi-shell approach: Multi-shell multi-tissue constrained spherical deconvolution — MRtrix 3.0 documentation
Hello, follow up on this from a year ago now that I have some participant data. So the original MAPMRI paper states that the model allows for the calculation of ODFs and SH’s from these ODFs. Is there any issue with using the ODFs/SH calculated using the dipy scripts (mapmri.py) as opposed to using the FODs/SH files calculated using the MRtrix line for tckgen? For tckgen I would be using the iFOD2 algorithm, where it used the FODs SH representation is used. Is there some processing within iFOD2 i should be aware of, for example if I provide an image file that is already in SH representation.
In summary, what I want to accomplish is implementing ACT Tractography on odfs/SHs derived from the MAPmri processing pipeline.
Any insight is much appreciated happy to clarify any points! Thank you for the time.
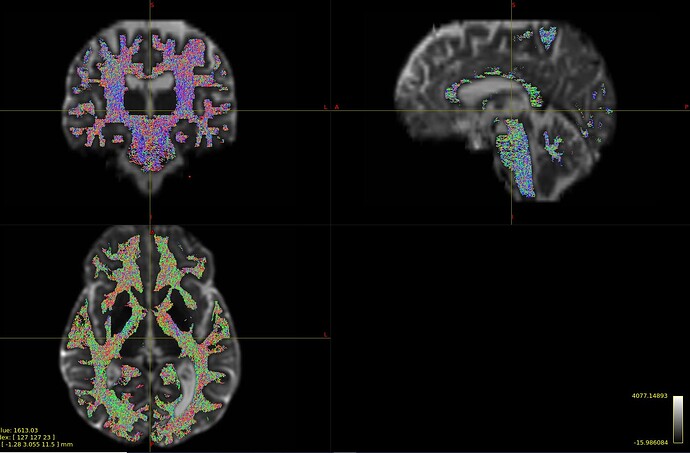
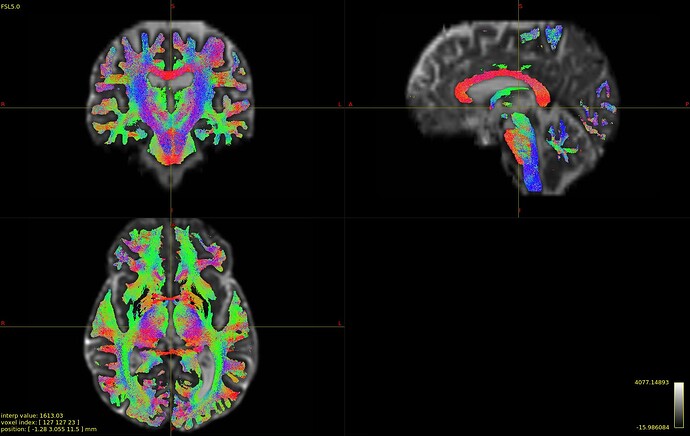
Please view the images of the resultant tractography below.
The mrtrix derived odfs produce more organized tract propagations compared to those odfs derived via the MAPmri.py processing pipeline. I thought this would help clarify what I was talking about in my previous reply. All parameters were kept the same, tractography was performed until 5m streamlines were obtained, sift (respective odf/fods were used for this as well) was applied to both to produce these tractograms of 1m streamlines. Any insight would be much appreciated.
Figure A. MAP-mri derived odf tractography. shconv was used to produce the SH representation of the odfs which was then used as the input for the tckgen function with iFOD2 as the algorithm. ODF calculations was restricted to the WM of the whole brain in order to provide some consistency with the MRtrix3 input for tckgen.
Figure B. Standard preprocessing and odf derivation via the dwi2fod function in MRtrix3. WM fods were used as the input for the tckgen function with the iFOD2 as the algorithm.