Hi,
I am trying to understand how exactly it is that MRTrix creates tracts directly from maps like FA instead of the raw diffusion data and what options would be best to carry this forward. Also, if anyone has a detailed understanding of how tract segmentation from model maps, I’d greatly appreciate that explanation too.
So far, I’ve been using this command: tckgen
-algorithm FACT -seed image T1 brain.nl
ii.gz dti_ FA.nii tracks dti.tck
I’d be open to feedback on this. [gene
rating tracts from model maps i.e.]
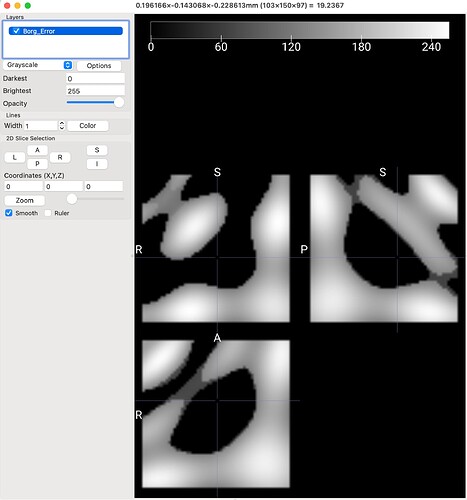
[a side question I had was viewing the .tck data generated…I don’t know if there is a scaling adjustment to be made or what but this is what I am getting when viewing in MRICroGL]
thank you!