Hi I am new to mrtrix and going through the tutorials on their website
http://jdtournier.github.io/mrtrix-0.2/tractography/preprocess.html
e.g. tensor components
I coped exactly as it says
entered
"dwi2tensor dwi.mif encoding.b dt.mif"
but it keeps giving this error.
dwi2tensor: [ERROR] expected exactly 2 arguments (3 supplied)
why is that?
Thank You
OK, first thing is that the website you’re referring to relates to the old 0.2 version of MRtrix, not MRtrix3. If you are using MRtrix3, the webpage you’re looking for is at http://userdocs.mrtrix.org/. If you are genuinely using MRtrix 0.2, then the support forum for this via the mrtrix-discussion mailing list - this forum is for the current MRtrix3 version. Note that the old 0.2 version should be considered obsolete, and has not been updated for quite a while…
That said, the dwi2tensor command has the same syntax in both versions. The command you’re providing is not what’s shown in the documentation: the encoding.b is either not required (if you’ve converted to dwi.mif directly from DICOM), or should be given using the -grad encoding.b option. You’re trying to provide encoding.b as an argument, which is why the command didn’t know what to do with it. As is stated in the doc, the correct command is either:
$ dwi2tensor dwi.mif dt.mif
or if the dwi.mif didn’t already contain the DW encoding:
$ dwi2tensor dwi.mif -grad encoding.b dt.mif
Note the -grad in the latter, it’s important…
Hi Mate I tried both commands you suggested and both gave errors attached below.
I am using mrtrix3 btw
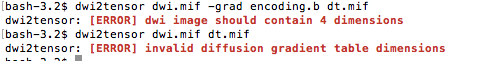
OK, looks like your DWI image isn’t as expected. What does mrinfo dwi.mif return? Your image should be 4D (3 spatial axes + volume axis)…
the original image was niftii which i converted to mrtrix format .mif
That doesn’t look like a DWI image: it’s a single 3D volume. It might conceivably be one volume from your DWI series, but there’s no way that constitutes a full DWI image. You’ll need to double-check where that image came from. Generally, I recommend you convert from DICOM directly to mif using mrconvert, that should take care of everything for you.
1 Like
How should I know if its a dwi image and not a single volume? What should it say?
In the mrinfo output you provided, you should look at the Dimensions field. Your current output reads 60 x 152 x 104, which basically tells you it’s a 3D image with those dimensions. A complete DWI dataset would consist of multiple volumes, and therefore be a 4D image. In that case, the Dimensions field should have 4 numbers; e.g., 60 x 152 x 104 x 65 for a DWI dataset that has 65 volumes.
As Donald suggested, it’s best to directly convert from your original DICOM images/folder to a mif image; so chances of losing data in the process are kept to a minimum. In your case, it’s very likely the NIfTI image was already only a single volume to begin with; so you lost your other volumes somewhere in an earlier step before that…
2 Likes

