Dear experts,
I am running FOD estimation on my 2-shell data (b=0 and b=1000). To my knowledge, the MSMT-CSD algorithm can be used for this purpose, but FOD estimation would be limited to 2 types of tissue with 2 b-values.
My question is, apart from WM, should I include the response function of GM or CSF for dwi2fod? Thanks a lot!!
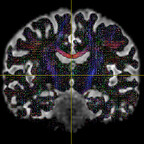
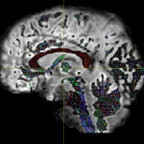
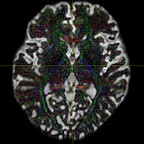
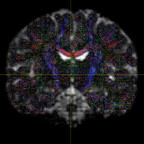
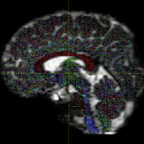
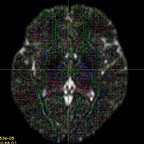
It has been suggested that, in order to fit all signal, the inputs typically have to be WM and CSF as these two have the lowest and highest diffusivities. However, I have tried both WM+GM and WM+CSF, and the WM+GM result seems to look more reasonable - it appears to be fitted on the WM region more accurately (i.e. with less false positive signals on non-WM regions):
WM+GM:
WM+CSF:
Please also let me know if it is suggestive of potential issues in the upstream processes. Many thanks!!