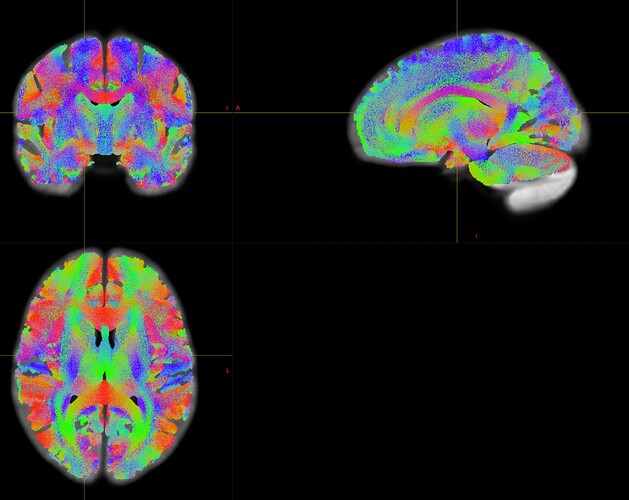
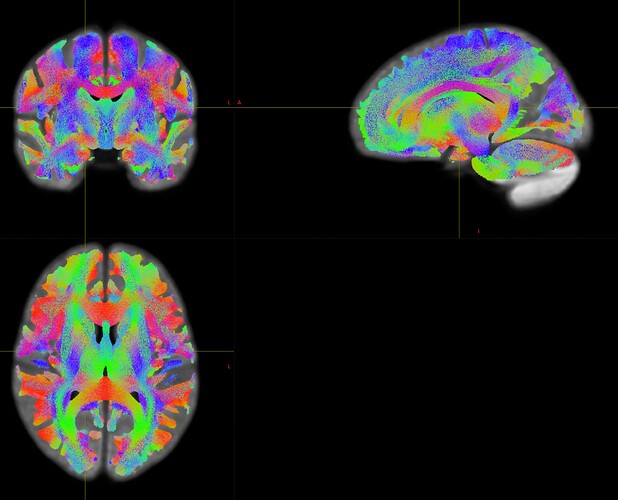
Dear MRtrix experts,
I was wondering if it is, in principle, possible to perform multicenter fixel-based analysis? And if yes, do I need to perform some extra steps?
I am working on a dataset of children at high-familial risk of developing severe mental illness.
Participants were scanned at two different sites with respectively a Siemens Prisma fit (64-channel head coil) or Skyra scanner (32-channel head coil). See below for some details on preprocessing, wm response functions retrieved for Prisma and Skyra data, and DWI sequence parameters.
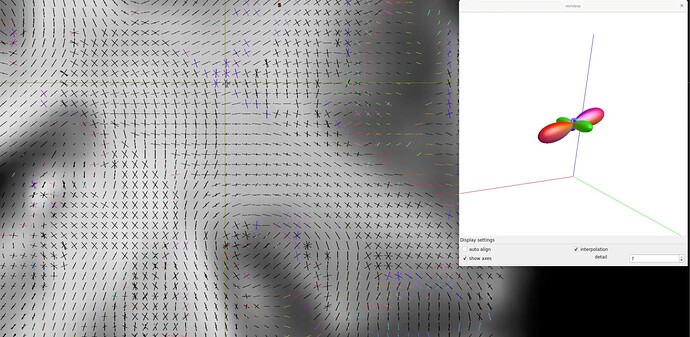
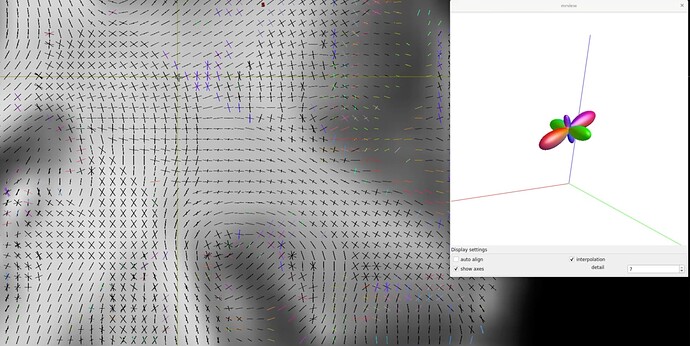
I notice that the first 4 Prisma values are approximately a factor 3 larger than those for Skyra.
I am looking forward to your response
Best wishes
William
Data were preprocessed with qsiprep1.6 :
- Denoising consisted of four steps:
- MP-PCA (Marchenko-Pastur distribution – Principal Component Analysis) denoising, as implemented in MRtrix3’s dwidenoise (Veraart et al., 2016), using a 5-voxel window.
- Gibbs unringing using Mrtrix3’s mrdegibbs (Kellner et al., 2016).
- B1 field inhomogeneity correction using dwibiascorrect from Mrtrix3 with the N4 algorithm (Tustison et al., 2010).
- b=0 image-based intensity normalization
- Head motion, eddy current, and susceptibility by movement distortion correction
- b=0 reference image creation
- Rigid 6-parameter (rotations and translations only) co-registration to the skull-stripped T1w image and DWI image resampling (1 mm isotropic) and gradient rotation.
I generated (and checked the fit) DWI masks using synthstrip-singularity (-b -2) on generated FA images
I used dhollander to retrieve wm, gm, and csf response functions:
dwi2response dhollander ${indata} \
-fslgrad ${inbvec} ${inbval} \
${OUT_DIR}/${bids_id}_dwi_${res}_response_wm.txt \
${OUT_DIR}/${bids_id}_dwi_${res}_response_gm.txt \
${OUT_DIR}/${bids_id}_dwi_${res}_response_csf.txt \
-mask ${newmask} \
-voxels ${bids_id}_dwi_${res}_response_voxel_selection.nii.gz \
-info \
-force
Example: wm response functions:
Prisma:
Shells: 5,1000
command_history: /home/wimb/anaconda3/envs/mrtrix3/bin/amp2response dwi.mif voxels_sfwm.mif safe_vecs.mif response_sfwm.txt -shells ‘5,1000’ (version=3.0.4)
21307.7996795693 0 0 0 0 0
11488.0266075202 -4509.69162685159 978.658571954375 -137.330096590635 11.2965465825901 4.40098708965145
skyra:
Shells: 5,999
command_history: /home/wimb/anaconda3/envs/mrtrix3/bin/amp2response dwi.mif voxels_sfwm.mif safe_vecs.mif response_sfwm.txt -shells ‘5,1000’ (version=3.0.4)
7053.85982998796 0 0 0 0 0
3723.35267227721 -1498.52515490046 328.62205129044 -54.0957827205385 7.7072439141182 6.4925266058699
DWI sequence parameters:
Prisma:
A diffusion-weighted imaging (DWI) sequence was setup for diffusion tensor imaging (DTI) and tractography and diffusion kurtosis imaging (DKI), using a pulse gradient spin echo (PGSE) echo planar imaging (EPI) sequence with uniformly distributed gradient directions. The DTI-tractography part consisted of 14 non-DW images (b = 0) and 80 DW images (b=1000 s/mm2); TR = 2541 ms; TE = 64 ms; flip angle = 90 degrees; matrix = 112 x 114; FOV = 200 x 204; phase partial Fourier = 6/8; echo spacing = 0.73 ms; bandwidth = 1540 Hz/Px; phase encoding = Anterior > Posterior, 75 transversal slices, multiband acceleration factor = 3; GRAPPA: acceleration factor = 2, reference lines = 24; 1.8 x 1.8 x 1.8 mm3 voxels.
skyra:
Diffusion-weighted imaging (DWI) consisted of two sequences. One for diffusion tensor imaging (DTI) and tractography and one for diffusion kurtosis imaging (DKI), both using a pulse gradient spin echo (PGSE) echo planar imaging (EPI) sequence with uniformly distributed gradient directions. The DTI-tractography part consisted of 13 non-DW images (b = 0) and 57 DW images (b=1000 s/mm2); TR = 2656 ms; TE = 77.2 ms; flip angle = 90 degrees; matrix = 100 x 100; FOV = 200 x 200; phase partial Fourier = 6/8; echo spacing = 0.68 ms; bandwith = 1665 Hz/Px; phase encoding = Anterior > Posterior, 72 transversal slices, multiband acceleration factor = 3; GRAPPA: acceleration factor = 2, reference lines = 24; 2 x 2 x 2 mm3 voxels