Hi.
Does anyone know how to perform correct skull stripping on neonatal T1w images?
I have tried with bet, bet2 and ROBEX with poor results.
Thanks,
Rosella
Hi,
One of the best algorithms I found for this data is MASS:
Best regards,
Manuel
Thank you very much. Do you know if it is implemented in a toolbox?
Best,
Rosella
Hi,
You can download it and install it from here: https://www.nitrc.org/projects/cbica_mass/
Best regards,
Manuel
Thank you very much. I also tried with Brainsuite but it needs a lot of manual correction, even if it works better than BET.
Best,
Rosella
Hi,
We did a study about neonatal skull-stripping: https://www.nature.com/articles/srep23470
I’m sorry, but I don’t have the code presented in the paper.
Best regards,
Manuel
thank you. Do you think mass algorithm can work?
rosella
Hi,
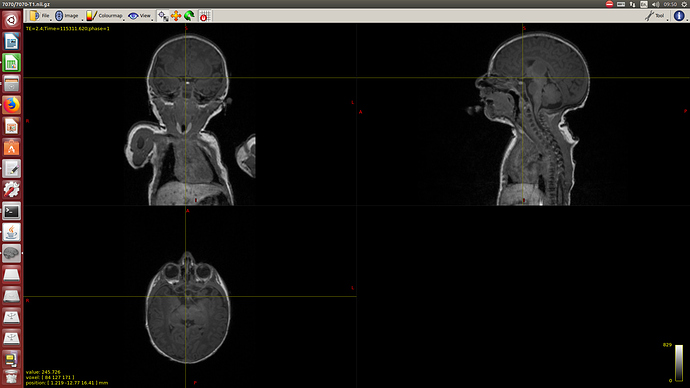
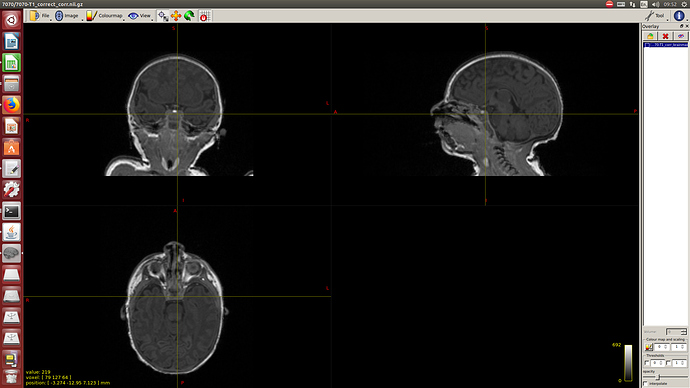
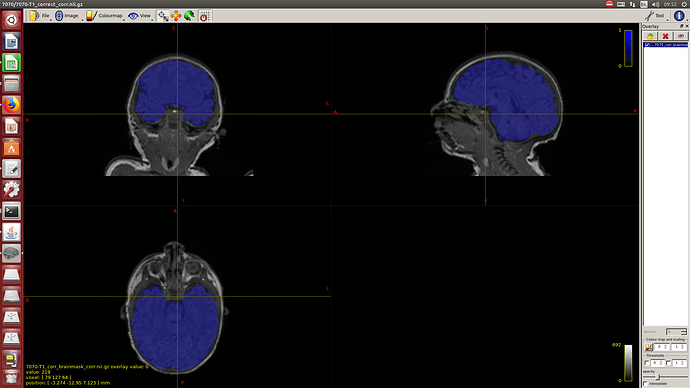
Is the one I’m currently using. Look here and example:
Raw data:
Data after running the full code for skull-stripping and bias correction:
And the mask:
I didn’t perform any manual corrections. If you are interested, this is the code I use:
robustfov -i ID-T1.nii.gz -r ID_FOVnormalized.nii.gz -b 130
bet ID_FOVnormalized.nii.gz ID_FOVnormalized_brain.nii.gz -R -m -f 0.3
N4BiasFieldCorrection -d 3 -i ID_FOVnormalized.nii.gz -x ID_FOVnormalized_brain_mask.nii.gz -o [ID-T1_corr.nii.gz,ID-T1_bias.nii.gz] -c [50x50x50,0.001] -s 2 -b [100,3] -t [0.15,0.01,200] -v 1
mass -in /path/ID-T1_corr.nii.gz -ref /path/templates/ -dest /path/ -NOQ -MT 6 -v
mrtransform ID-T1_corr_brainmask.nii.gz -replace ID_FOVnormalized.nii.gz ID-T1_corr_brainmask_corr.nii.gz
N4BiasFieldCorrection -d 3 -i ID_FOVnormalized.nii.gz -x ID-T1_corr_brainmask_corr.nii.gz -o [ID-T1_correct_corr.nii.gz,ID-T1_corr_bias.nii.gz] -c [50x50x50,0.001] -s 2 -b [100,3] -t [0.15,0.01,200] -v 1
Note that MASS requires a templates folder, you could use the subjects of the dHCP to create that folder. I hope this helps.
Best regards,
Manuel
Ok I will try in order to avoid manual revision.
thanks,
Rosella
One more question, I need to work with DKI data and I will preprocess them with the Mrtrix3 pipeline I think. However, as regards the 3dT1 image that I use as an anatomical reference for registration to atlas and parcellization, does it need any processing in your opinion?
thanks,
Rosella
Hi,
I would say than having a good mask is very important for the atlas registration to the T1w. The bias field correction would have less impact for the registration (I think). The mask it also would affect for the registration of the T1w and the B0.
Best regards,
Manuel
So do you suggest a bias correction step and that’s it?
Rosella
Hi,
Yes, that’s the only thing I usually do in the T1w.
Best regards,
Manuel
Thank you very much for your answers. And as regards diffusion images, do you think the diffusion preprocessing pipeline developed by Mrtrix3 is suitable also for DKI neonatal images?
Thanks
Rosella
Hi,
I’m not familiar with DKI data, but I would say that as long as the acquisition gradients are distributed in a shell (or multi-shell), everything should work.
Best regards,
Manuel
I am finding difficulties in installing MASS on linux laptop. In particular as regards setting the SCHEDULER option. Do you remember having any problems of this kind?
thanks ,
Rosella
Hi,
I don’t remember any error, I just run everything with default (after installing all the other dependencies)
Best regards,
Manuel
I installed the dependencies but it fails at testing step, after building:
rosella@vivi:~/Downloads/mass-1.1.0-build$ make test
Running tests…
Test project /home/rosella/Downloads/mass-1.1.0-build
Start 1: mass_fillholes_test
1/8 Test #1: mass_fillholes_test …***Failed 0.03 sec
Start 2: mass_morpho-dilate_test
2/8 Test #2: mass_morpho-dilate_test …***Failed 0.03 sec
Start 3: mass_morpho-erode_test
3/8 Test #3: mass_morpho-erode_test …***Failed 0.02 sec
Start 4: mass_chooseTemplates_test
4/8 Test #4: mass_chooseTemplates_test …***Failed 0.02 sec
Start 5: mass_registrations_test
5/8 Test #5: mass_registrations_test …***Failed 0.04 sec
Start 6: mass_skullstripping_test
6/8 Test #6: mass_skullstripping_test …***Failed 0.02 sec
Start 7: mass_thresholdJacobian_test
7/8 Test #7: mass_thresholdJacobian_test …***Failed 0.03 sec
Start 8: mass_system_test
8/8 Test #8: mass_system_test …***Failed 0.03 sec
0% tests passed, 8 tests failed out of 8
Label Time Summary:
mass = 0.21 sec
Total Test time (real) = 0.24 sec
The following tests FAILED:
1 - mass_fillholes_test (Failed)
2 - mass_morpho-dilate_test (Failed)
3 - mass_morpho-erode_test (Failed)
4 - mass_chooseTemplates_test (Failed)
5 - mass_registrations_test (Failed)
6 - mass_skullstripping_test (Failed)
7 - mass_thresholdJacobian_test (Failed)
8 - mass_system_test (Failed)
Errors while running CTest
Makefile:137: recipe for target ‘test’ failed
make: *** [test] Error 8
Hi,
This happened also to me, I think this is a path problem (something I didn’t choose correctly during the configuration process). The test failed but then the softare was installed and running without errors. Could you run make install and then go to the path/mass/bin folder and run ./mass
If is the same case as me, it should work.
Best regards,
Manuel
Sorry, just one more thing. I noticed MASS and ANTs require 2 different verions of cmake which are not compatible between each other. Is there a way to maintain the 2 versions?
thanks,
Rosella