Hello
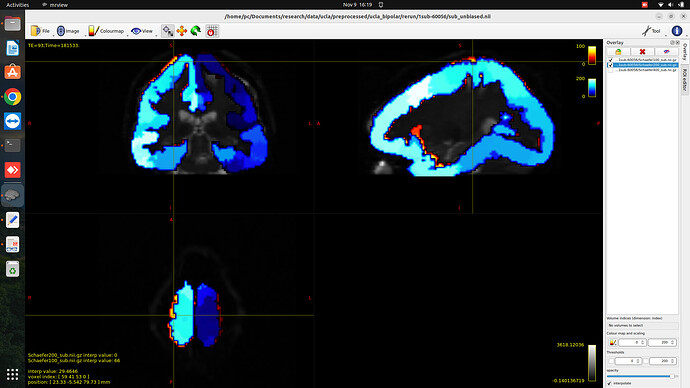
WHen i am trying to coregister parcellation images (say, Schaefer200 and Schaefer400) to DTI space for particular subject, registration is good for one parcellation (say Schaefer200) and not gooed for the other. I am follwing all the same steps for preprocessing. but for Schaefer400 registration is poor.
Could anyone give any suggestions, why this happening
1 Like
What are the exact commands you are using? Without this, it will be very hard for anyone to help.
flirt -in $FSL_DIR/data/standard/MNI152_T1_1mm_brain.nii.gz -ref mean_b0_masked -omat mni2dti.mat -datatype int -out mni2dti.nii.gz
fnirt --in=$FSL_DIR/data/standard/MNI152_T1_1mm_brain.nii.gz --ref=mean_b0_masked.nii.gz --aff=mni2dti.mat --cout=mni2dti_transf #–config=T1_2_MNI152_2mm.cnf
applywarp --ref=mean_b0_masked --in/home/pc/…/Schaefer2018_LocalGlobal/Parcellations/MNI/Schaefer2018_200Parcels_7Networks_order_FSLMNI152_1mm.nii.gz --warp=mni2dti_transf --out=Schaefer200_sub.nii.gz --interp=nn
#applywarp --ref=mean_b0_masked --in=/home/pc/…/Schaefer2018_LocalGlobal/Parcellations/MNI/Schaefer2018_400Parcels_7Networks_order_FSLMNI152_1mm.nii.gz --warp=mni2dti_transf --out=Schaefer400_sub.nii.gz --interp=nn
Step 5. - Generating connectome
tck2connectome -symmetric -zero_diagonal -scale_invnodevol -tck_weights_in sift_1M.txt tracks_10M.tck Schaefer200_sub.nii.gz SC_200.csv -out_assignment assignments_SC200.csv
Step 5. - Generating connectome
tck2connectome -symmetric -zero_diagonal -scale_invnodevol -tck_weights_in sift_1M.txt tracks_10M.tck Schaefer400_sub.nii.gz SC_400.csv -out_assignment assignments_SC400.csv
these are the commands i used.
You are using volumetric ROIs for those parcellations, which will definitely result in two issues:
- Overlapped/intermixed ROIs, even though the interpolation is nearest neighbour
- Poor fiber to ROI assignment in your connectome step.
Obviously, these issues are more apparent when you do include higher granularity for your parcellation. The best solution maybe to map any parcellation you have using FreeSurfer commands on your recon-all-ed native T1s “surface” (, and then apply transforms you have for T1-to-DWI registration.
Check the guidelines here:
Amir
Thank you Amir.
I follow the steps
mri_surf2surf --hemi lh
–srcsubject fsaverage
–trgsubject sub_T1w_recon
–sval-annot /home/pc/Documents/research/percallation_file/Schaefer2018_LocalGlobal/Parcellations/FreeSurfer5.3/fsaverage/label/rh.Schaefer2018_200Parcels_7Networks_order.annot
–tval $SUBJECTS_DIR/sub_T1w_recon/label/lh.Schaefer2018_200Parcels_7Networks_order.annot
The commands generate a error ‘Permision denied’ and file is not generated.
*Setting mapmethod to nnf
Id: mri_surf2surf.c,v 1.103 2015/11/05 22:07:33 greve Exp
setenv SUBJECTS_DIR /usr/local/freesurfer/subjects
cd /home/pc/Documents/research/data/test_Scheafer
mri_surf2surf --hemi lh --srcsubject fsaverage --trgsubject sub_T1w_recon --sval-annot /home/pc/Documents/research/percallation_file/Schaefer2018_LocalGlobal/Parcellations/FreeSurfer5.3/fsaverage/label/rh.Schaefer2018_200Parcels_7Networks_order.annot --tval /usr/local/freesurfer/subjects/sub_T1w_recon/label/lh.Schaefer2018_200Parcels_7Networks_order.annot
sysname Linux
hostname pc-HP-Pavilion-Laptop-14-dv1xxx
machine x86_64
user pc
srcsubject = fsaverage
srcval = (null)
srctype =
trgsubject = sub_T1w_recon
trgval = /usr/local/freesurfer/subjects/sub_T1w_recon/label/lh.Schaefer2018_200Parcels_7Networks_order.annot
trgtype =
srcsurfreg = sphere.reg
trgsurfreg = sphere.reg
srchemi = lh
trghemi = lh
frame = 0
fwhm-in = 0
fwhm-out = 0
label-src = (null)
label-trg = (null)
OKToRevFaceOrder = 1
UseDualHemi = 0
Reading source surface reg /usr/local/freesurfer/subjects/fsaverage/surf/lh.sphere.reg
Loading source data
Reading surface file /usr/local/freesurfer/subjects/fsaverage/surf/lh.orig
reading colortable from annotation file…
colortable with 101 entries read (originally Schaefer2018_200Parcels_7Networks)
Reading target surface reg /usr/local/freesurfer/subjects/sub_T1w_recon/surf/lh.sphere.reg
Done
Using surf2surf_nnfr()
Mapping Source Volume onto Source Subject Surface
surf2surf_nnfr: building source hash (res=16).
Surf2Surf: Forward Loop (151617)
Surf2Surf: Dividing by number of hits (151617)
INFO: nSrcLost = 47687
nTrg121 = 151617, nTrgMulti = 0, MnTrgMultiHits = 0
nSrc121 = 88318, nSrcLost = 47687, nSrcMulti = 75524, MnSrcMultiHits = 0.838131
Saving target data
Converting to target annot
Saving to target annot /usr/local/freesurfer/subjects/sub_T1w_recon/label/lh.Schaefer2018_200Parcels_7Networks_order.annot
could not write annot file /usr/local/freesurfer/subjects/sub_T1w_recon/label/lh.Schaefer2018_200Parcels_7Networks_order.annot
Permission denied
*
Is the problem with freesurfer?
I generally suggest not to set SUBJECTS_DIR inside root subdirectories such as /usr/local. I think that’s the problem, and requires super user credentials (sudo mri_surf2surf …), which is not ideal in general.
Amir
Thank you Amir.
I reinstall freesurfer and set the path to home folder. then the steps run fine.
Hello Amir,
I need to add some subcortical regions in Schaefer200 parcellation. How can i use subcortical regions to these parcellation file? Can i do it using the same process you told me initially.
(https://github.com/ThomasYeoLab/CBIG/tree/master/stable_projects/brain_parcellation/Schaefer2018_LocalGlobal/Parcellations/MNI)
For subcorticals, you can use the first pipeline you had: using inverse t1-to-mni warps to bring subcortical ROIs into native space. In my experience, ANTs outweighed all other tools to bring subcorticals into native. I think you can also find some information about it in BATMAN guideline on MRtrix.
Amir